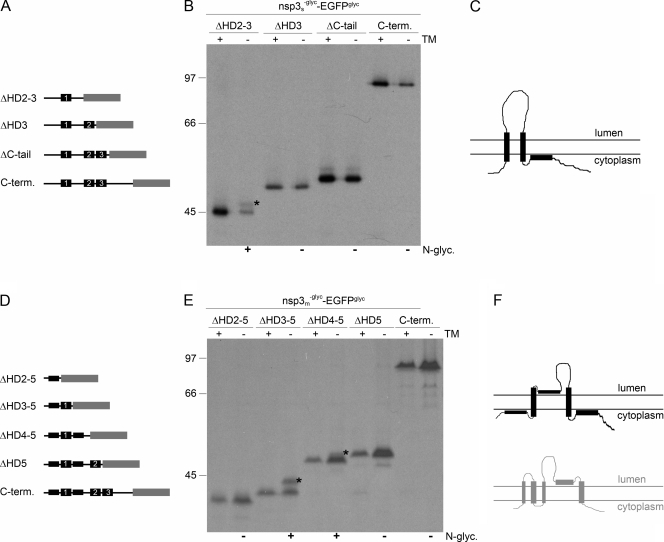
FIG. 5.
Membrane integration of deletion mutant forms of SARS-CoV and MHV nsp3. (A, D) Schematic representations of the C-terminal (C-term.) deletion mutant forms of SARS-CoV (A) and MHV (D) nsp3, with the hydrophobic domains presented as black rectangles and the EGFPglyc tag in gray. The corresponding hydrophobic domains in the two proteins are indicated by numbers. (B, E) vTF7-3-infected OST7-1 cells were transfected with the indicated constructs and expressed in the presence (+) or absence (−) of tunicamycin (TM). The cells were labeled with [35S]methionine from 5 to 6 h p.i., lysed, and processed for immunoprecipitation with anti-EGFP antiserum followed by SDS-PAGE. The positions and masses (in kilodaltons) of the protein size markers are indicated at the left. The asterisks indicate the position of the glycosylated protein species. Below the panels, the observed presence or absence of N glycosylation is indicated by a plus or a minus sign, respectively. (C, F) Models of the membrane structures of SARS-CoV (C) and MHV (F) nsp3, with the hydrophobic domains presented as black rectangles. For comparison, the MHV nsp3 model proposed by Baker and coworkers is shown below in gray.