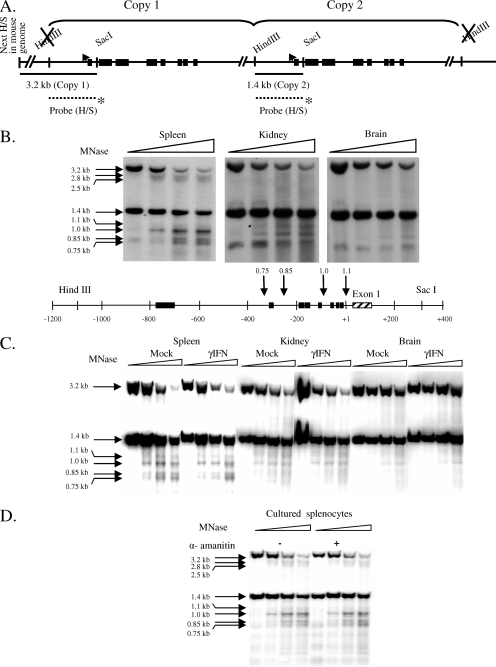
FIG. 5.
MNase hypersensitivity correlates with expression level in tissues but does not change with induction or depend on active transcription. (A) Map of the PD1 gene in B10.PD1 transgenic mice. The regions examined for nuclease-hypersensitive regions are indicated by horizontal lines. The labeled HindIII/SacI probe is represented by a dotted line with an asterisk at the bottom of the figure. Its location shows where it hybridizes to the PD1 gene. (B to D) Southern blots with MNase-treated and HindIII/SacI-digested samples from untreated (B) and mock- and IFN-γ-treated spleen, kidney, and brain (C) or from splenocytes cultured in medium alone or medium with α-amanitin (D) probed with a fragment corresponding to the 1.4-kb PD1 5′ region are presented. Fragment sizes corresponding to the band are marked on the left. At the bottom of panel B, arrows indicate the positions of MNase-hypersensitive sites at the 5′ end of the PD1 gene, and the numbers on top of the arrows represent sizes of the bands (in kilobases) generated as a result of MNase hypersensitivity. Note that the MNase concentration was standardized to optimize the cleavage of the 1.4-kb fragment and the generation of its derivative 1.0-kb hypersensitive band. These conditions of MNase result in greater digestion of the 3.2-kb fragment (upstream copy of the gene). Due to the decreased MNase hypersensitivity in the kidney and brain, the weaker hypersensitive bands derived from the 3.2-kb fragment are not always visible.