Figure 2.
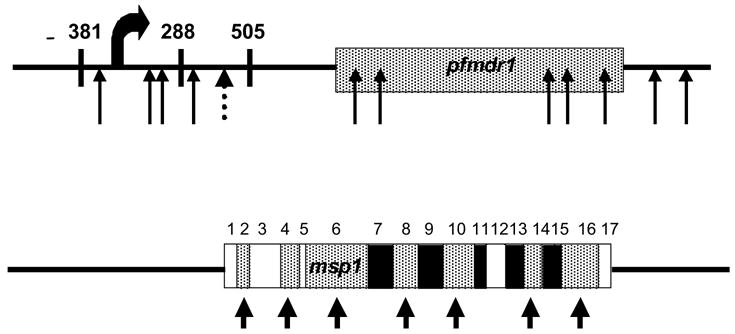
SNPs are Nonrandomly Distributed Between Coding and Noncoding Regions of a Gene. The pfmdr1 gene (PFE1150w; coding region from position 957,885 – 962,144; shown from position 955,110 – 962,912) contains SNPs both within the coding region (arrows corresponding to Tyr86, Phe184, Cys1034, Asp1042, Tyr1246) and in the 5′ (2775 base pairs (bp)) and 3′ (768 bp) flanking regions. One of these SNPs (dotted arrow) is present in both field and laboratory isolates. Several of these mutations may be functionally relevant due to the proximity to the start of transcription (curved arrow). The msp1 gene (PFI1475w; coding region from position 1,201,802 – 1,206,964; shown from position 1,200,803 – 1,208,021) contains several SNPs within the coding region (represented by arrows) within the highly polymorphic blocks (2, 4, 6, 8, 10, 14, 16) defined by Tanabe and colleagues (Miller et al., 1983) shown as conserved (open), semiconserved (hatched), and variable (solid) regions. Most of this variation is dimorphic and grouped into the allelic groups of K1-type and MAD20-type alleles. Exception to this dimorphism is present in Block 2 with additional degenerate tripeptide repeats that represent the RO33-type allele. Both the 5′ (999 bp) and the 3′ (1057 bp) flanking regions are monomorphic among eight or more genetically diverse parasites and lack SNPs in the dataset analyzed.
