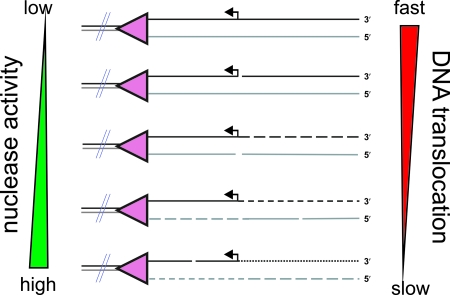
FIG. 5.
Effect of ATP and Mg2+ ions on the cleavage pattern observed upon DNA processing by RecBCD. During DNA translocation and unwinding, RecBCD cleaves both nascent single strands using a single nuclease active site. The frequency of cleavage on each strand (i.e., the average distance between cleavage points) is dependent on the voracity of the nuclease domain (which increases with increasing free Mg2+ ion concentrations) and the translocation rate (which increases with increasing ATP:Mg2+ concentrations) as indicated. For example, a fast-moving enzyme with low nuclease activity will produce mostly full-length ssDNA (top DNA molecule shown), whereas a slow-moving enzyme with high nuclease activity will produce very closely spaced cleavage points in the unwound DNA (last DNA molecule shown). The relative accessibility of each strand to the nuclease domain is also an important factor, and this is controlled by Chi recognition. Consequently, RecBCD can produce a spectrum of cleavage products dependent upon the precise in vitro solution conditions. Under physiological conditions (1 to 3 mM ATP [39] and 1 to 2 mM free magnesium ion [2]), the processing of Chi-containing DNA by RecBCD results largely in products similar to those depicted in the third and fourth DNA molecules illustrated.