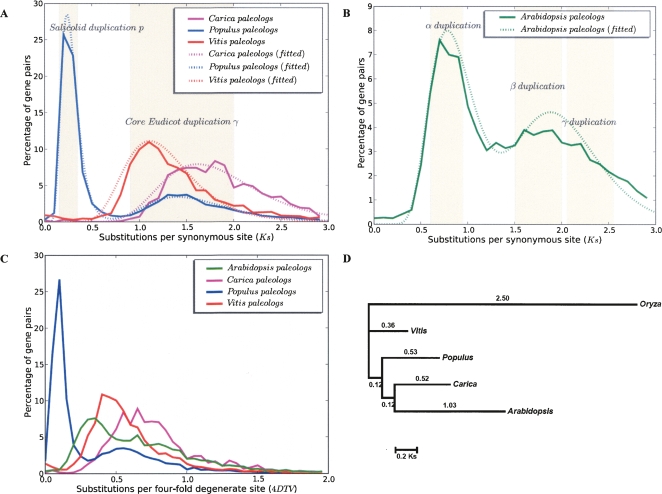
Figure 4.
(A,B) Distribution of Ks distances among Carica, Populus, Vitis, and Arabidopsis paleologs. Ks values are grouped into bins of 0.1 intervals. Certain Ks intervals are highlighted as they correspond to several presumed whole-genome duplication events. Dotted lines are fitted mixtures of log-normal distributions for the paleolog Ks distributions (see Methods). (C) Distribution of 4DTV distance among paleologs in the same four eudicot lineages. (D) Phylogeny of single-copy ortholog set used in relative rate estimates. A total of 47 orthologous genes that are single copy in all five species were used in the analysis. Protein alignments for each ortholog group were constructed and then used to guide DNA alignments. The alignments are then concatenated, with 53,856 aligned nucleotide positions. Per-site Ks values on each branch were estimated by codeml in the PAML package (Yang 1997) using a constrained topology that reflects organismal relationships.