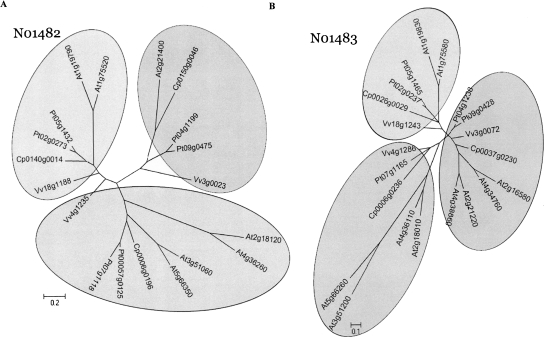
Figure 5.
Phylogenetic analysis of ancestral loci N01482 (A) and N01483 (B). Coding sequences of all members in four eudicot species for each ancestral locus (19 genes in N01482, 21 in N01483) were aligned by CLUSTALW (Thompson et al. 1994) using parameters suggested by Hall (2007). Phylogenetic relationships among the members and sequences were grouped into clades using MrBayes (Ronquist and Huelsenbeck 2003). The Bayesian analysis was carried out for 500,000 generations using the General Time Reversible plus Gamma (GTR+G) substitution model selected based on MODELTEST (Posada and Crandall 1998). All branches with support <50% are collapsed into a polytomy. A majority tree was presented in both cases. The gene names for Carica, Populus, and Vitis are recoded to reflect relative orders on chromosome or scaffold (see Methods). The conversions from the original locus identifiers to the re-indexed gene names are available as a conversion table in Supplemental Data 4. In case the original gene identifiers are subject to future changes, the conversion table will be updated accordingly. Arabidopsis gene names follow their standard TAIR locus IDs. Scale bars represent the number of substitutions per site following the GTR+G model.