Figure 3.
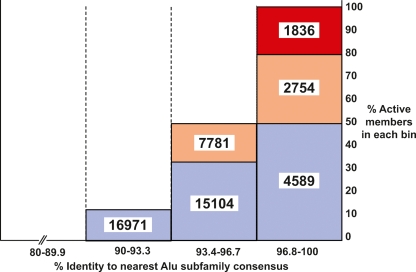
How many potentially active Alu core elements in the human genome? A model was developed for estimating the number of potentially active Alu core elements in the reference human genome. A set of 33 unbiased Alu copies (see Methods) were placed into four bins according to their level of sequence variation (96.8%–100%, 93.4%–96.7%, 90%–93.3%, and <89.9%). The percentage of “active copies” was calculated for each bin, where “active” was defined as >5% of AluYa5 activity level in the mobilization assay. The percentages of active copies within each bin were then used to estimate how many genomic copies with the same levels of variation are present in the human genome (the results are depicted as numbers). The levels of activity were broken down further into “hot” (red; 100%–66.6% of AluYa5 activity level), “moderate” (yellow; 66.5%–40% of AluYa5), and “cool” (blue; 39.9%–5% of AluYa5). No elements below 90% conservation were active in the mobilization assay. This method provides a liberal estimate of the number of Alu core elements that would be active if cloned and tested in our mobilization assay. The actual number of elements expressed and mobilized from their natural genomic locations is likely to be lower than the numbers presented due to the impact of flanking genomic regions on Alu expression (see Discussion).