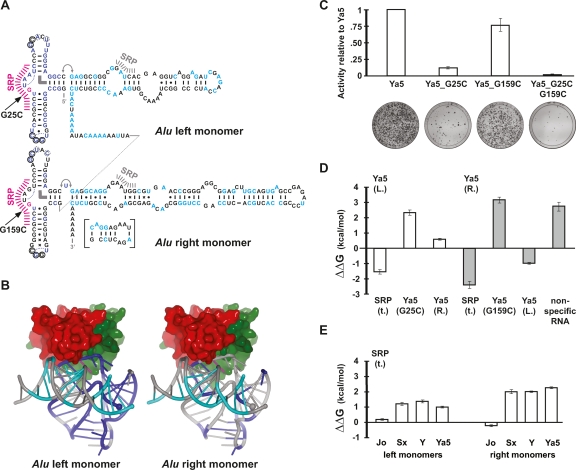
Figure 4.
SRP9/14 host proteins are necessary for efficient Alu retrotransposition. (A) Secondary structure representation of an AluYa5 RNA. The 124 positions (Supplemental Fig. 1) that are conserved among 70 consensus sequences and 45 experimentally tested, active Alu elements are highlighted in blue (Alu 5′ domain) and cyan (Alu 3′ domain). Positions (G25 and G159) that were mutated to prevent SRP9/14 binding are in magenta. Hash marks indicate major (magenta) and minor (gray) SRP9/14 contact sites. The inset shows an alternative base-pairing that is possible since the emergence of the AluS family and that may be responsible for the drop in SRP9/14 affinity at the transition from the AluJ to S families. Thin curves indicate U-turns, the thick, curved bar indicates a stacking interaction, and the double arrow a flexible linkage. Circles indicate tertiary base pairs between the loops. The dotted circles symbolize an alternative base pair of the respective nucleotides with G14 (left monomer) and G148 (right monomer). Additional symbols: (|) Watson-Crick base pairs; (.) wobble base pairs; (x) other (potential) base pairs. (B) Three-dimensional representation of Alu RNA and SRP9/14 binding. Positions highlighted in A have been mapped onto the crystal structure of the SRP Alu RNP (Weichenrieder et al. 2000) using the same colors. SRP9 is in red and SRP14 is in green. The 5′ and 3′ RNA ends are indicated by a large and small sphere, respectively. (C) Retrotransposition activity of consensus AluYa5 and SRP9/14 binding mutants. The mobilization activities relative to AluYa5 are shown along with representative assay plates below. (D) Relative affinities of Alu RNA mutants for SRP9/14. Left and right mutant Alu RNA monomers competed against the wild-type AluYa5 left monomer RNA (Ya5 (L.), white bars) or AluYa5 right monomer RNA (Ya5 (R.), gray bars) as labeled references in an in vitro assay based on nitrocellulose filter binding of SRP9/14. Positive values of ΔΔG reflect loss of affinity with respect to wild-type RNA. The full data set is presented in Supplemental Table 2. A representative binding experiment is shown in Supplemental Figure 2. (E) Relative affinities of Alu consensus sequences for SRP9/14. In contrast to D, truncated SRP RNA (SRP(t.)) was used as reference. These binding results agree with previous gel-shift assays examining SRP9/14 binding to AluS and Y monomers (Sarrowa et al. 1997).