Figure 1.
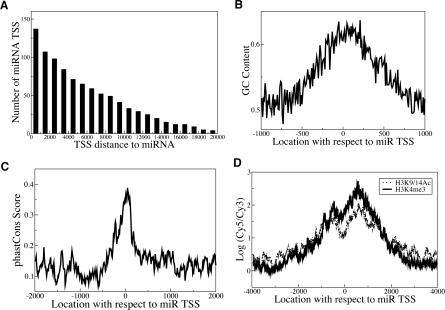
miRNA promoters exhibit similarities to known RNAPII promoters. (A) Cumulative histogram showing miRNA promoter distances relative to the corresponding mature miRNA locations in the human genome. The Y-axis represents the number of miRNA promoters with promoter-to-mature miRNA distance greater than or equal to the indicated x-value. (B,C) Average GC content (B) and evolutionary conservation scores (Siepel et al. 2005) (C) are shown at miRNA promoters. The high GC content and evolutionary conservation near transcription initiation regions are similar to the characteristic features found in coding genes (Ozsolak et al. 2007). (D) Composite H3K4me3 and H3K9/14Ac profiles at novel miRNA TSS regions. The X-axis represents the distance relative to the transcription initiation region, the Y-axis the average H3K4me3 or H3K9/14Ac ChIP–chip signal of all miRNA promoters at the indicated x-value. The concentrated H3K4me3 and H3K9/14Ac modifications around transcription initiation regions are similar to the patterns observed for coding gene TSSs (Heintzman et al. 2007).