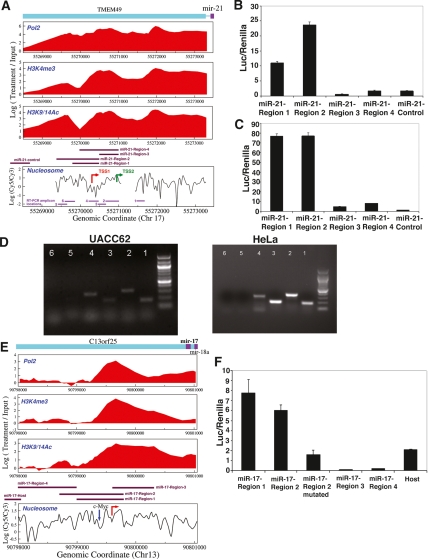
Figure 2.
Identification of miR-21 and miR-17-cluster transcription initiation regions. (A) A nucleosome-depleted area (TSS1) 617 bp upstream of the published position (TSS2) may be the miR-21 TSS. (B,C) To identify regions with promoter function, several segments shown in A were cloned upstream of a luciferase reporter. miR-21-Region-1,2,3,4 and miR-21-control pGL3-basic luciferase reporter constructs (A) (Supplemental Table S6) were transfected into UACC62 (B) and HeLa (C). While miR-21-Region-1 and Region-2 (corresponding to the novel promoter we identified) exhibited promoter activity in both cell lines, we detected weaker activity above negative control levels from region-3 and region-4 (corresponding to the published promoter position) in HeLa, but not in UACC62. Region-4 contained the novel transcription initiation region but not the upstream conserved TF-binding sites. The Y-axis shows the firefly luciferase activity normalized to the control renilla for each sample. (D) RT–PCR was performed using primers designed for the 3′ end of pri-miR-21 construct (Cai et al. 2004) for the RT step. PCR was performed with primer pairs in A (Supplemental Table S7). (E,F) miR-17-cluster genomic regions surrounding the novel (E, red arrow pointing toward right) and host TSSs (Supplemental Table S6) were cloned upstream of luciferase reporter and transfected into UACC62. Unlike negative control constructs (region-3 and region-4), Region-1, Region-2, and host exhibited luciferase activity (F). Mutation of c-Myc site (E, blue down arrow) in miR-17-Region-2 construct (miR-17-Region-2-mutated) resulted in an approximately threefold decrease in luciferase activity relative to the wild-type construct. Data presented are mean ± SEM from three independent experiments.