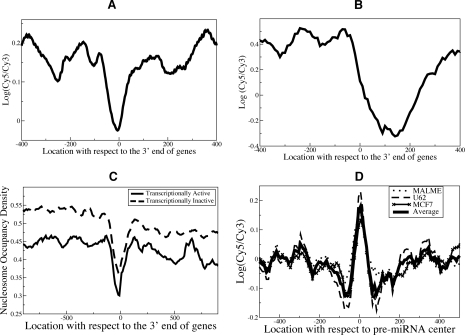
Figure 6.
Nucleosome positioning in genomic regions surrounding the 3′ end of genes and mature miRNAs-coding sequences exhibits distinct patterns. (A,B) Nucleosome positioning signals at the 3′ end of genes of RefSeq genes in HeLa (A) and translation end sites in yeast (Yuan et al. 2005) (B) were aligned, and average probe signals were calculated. (C) We analyzed the nucleosome positioning maps generated by another group for human CD4+ T cells (Schones et al. 2008) and observed a similar nucleosome depletion at the 3′ ends of transcriptionally active and inactive human genes, further validating our findings. Transcriptionally active genes have less nucleosome occupancy, which may be simply due to nucleosome remodeling/eviction during transcription. (D) Nucleosome positioning signals surrounding all pre-miRNAs were aligned at pre-miRNA centers, and average probe signals were calculated. Preferential occupancy of positioned nucleosomes in pre-miRNA regions (Supplemental Fig. S9C) was observed.