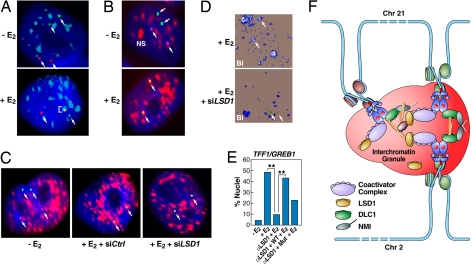
Fig. 5.
Interchromatin granules are hubs for interchromosomal interactions. (A) 2D-FISH shows selective association of interacting loci with interchromatin granules. Only the interacting (TFF1:GREB1) alleles intermingle within interchromatin granules (ICGs) stained with αSC35 (pseudocolored blue), whereas the remaining noncolocalized alleles show no colocalization with ICGs. (B) 2D-FISH of biallelic TFF1/GREB1 interactions (purple/green as indicated by arrows) coincident with the IGCs. (C) LSD1 is required for the association of the interacting TFF1:GREB1 loci with IGCs (pseudocolored red). Microinjection of siRNA against LSD1 abolished the colocalization between the interacting TFF1:GREB1 loci and ICGs. (D) 3D-FISH demonstrating the requirement of LSD1 for association of the interacting TFF1:GREB1 loci (arrows) with IGCs. (E) Percentage of cells exhibiting IGC association in response to control and specific siRNA against LSD1 (**, P < 0.001 by χ2). The rescure experiments indicate that the enzymatic activity of LSD1 is at least partially required for mediating the association of the interacting gene loci with ICGs. (F) Proposed model of E2-induced, actin/myosin1/DLC1-mediated chromosomal movement and LSD1-dependent interactions with interchromatin granules, creating a 3-dimensional enhancer hub in the nucleus.