Figure 1.
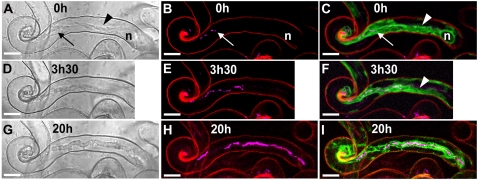
Cytoarchitecture associated with IT development in M. truncatula root hairs. Three successive stages of IT growth in a root hair of the supernodulating M. truncatula mutant sunn expressing the 35S-GFP-HDEL transgene (4 d postinoculation). Bright-field images are shown on the left (A, D, and G). The corresponding confocal images (z axis projections of serial optical sections) showing the cCFP fluorescence of the rhizobia (magenta) are presented in B, E, and H, and combined with the ER-targeted GFP-HDEL (green) in C, F, and I. Cell wall autofluorescence in the confocal images is in red. A to C, At this early stage of IT development, there are only a very few bacteria in the thread, and several gaps are clearly visible within the file (B). The broad ER-rich cytoplasmic bridge (A and C, arrowheads) connecting the nucleus (n) with the elongating IT (arrow) is characteristic of actively growing ITs (C). Note that the IT tip is embedded in the dense, ER-rich cytoplasm and therefore difficult to visualize in the bright-field image. D to F, 3.5 h later, the IT has progressed within the root hair (D), and the rhizobia multiplied within the thread (E). Gaps are again visible within the single file of bacteria. The more mature section of the IT is surrounded by a layer of cytoplasm, whereas the newly synthesized region is embedded in the cytoplasmic bridge (F, arrowhead). Note that the root hair nucleus has moved down the shaft of the root hair and is no longer visible in the image frame. G to I, 20 h later, the IT has grown further down the hair toward the base of the cell. The bright-field image (G) shows the clear outline and uneven surface of the typical mature IT, and the confocal image (I) reveals that this fully differentiated IT is still surrounded by a thin layer of ER-labeled cytoplasm. Thicker stretches of the colonizing bacterial file (H) suggest that there is a doubling in certain regions (H) and, even at this late stage, bacteria-free gaps are still present along the file. Bars = 10 μm.