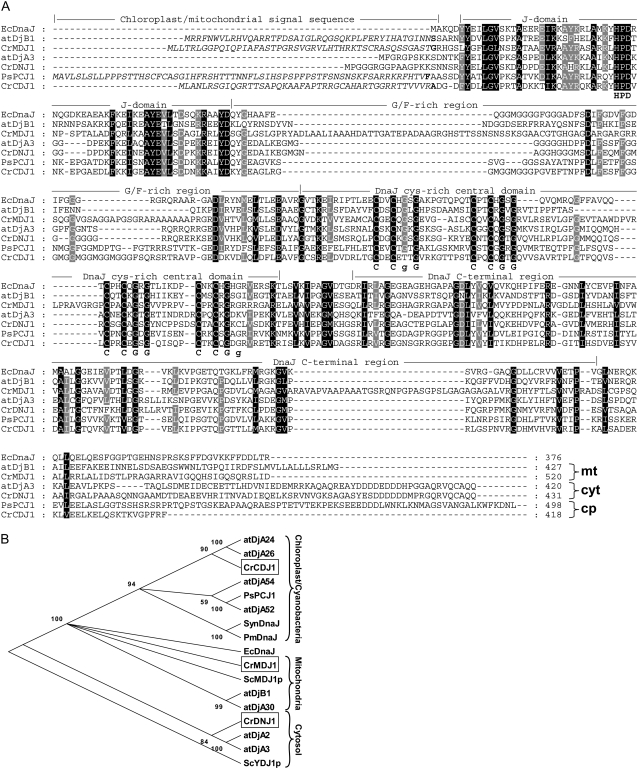
Figure 1.
Alignment and phylogenetic tree of DnaJ homologs. A, Alignment of amino acid sequences of DnaJ homologs. Sequences aligned are from E. coli DnaJ (EcDnaJ, accession AAA23693); from Arabidopsis mitochondrial and cytosolic DnaJ homologs (atDjB1 and atDjA3, accessions NP_174142 and AAB49030, respectively); from Chlamydomonas cytosolic, chloroplast, and mitochondrial DnaJ homologs (CrDNJ1, CrCDJ1, and CrMDJ1, accessions EDP04706, AAU06580, and gene model estExt_fgenesh1_pg.C_870033 at http://genome.jgi-psf.org/Chlre3/Chlre3.home.html, respectively); and from a pea chloroplast DnaJ homolog (PsPCJ1, accession CAA96305). Amino acids highlighted in black are conserved in all seven proteins; those highlighted in gray are conserved in at least six of them. Cys and Gly of the four conserved CXXCXGXG motifs are given below the sequences in bold letters. Italicized sequences represent organellar transit peptides as predicted by the TargetP program (Emanuelsson et al., 2000), with the residue in boldface corresponding to the first amino acid of the mature protein. Domain designations are according to Cheetham and Caplan (1998). B, Phylogram based on an alignment of the amino acid sequences of ZFD-containing J-domain proteins from Chlamydomonas, Arabidopsis, yeast, pea, E. coli, and two cyanobacteria. Sequences were from the same sources as in A, but also included additional sequences from Arabidopsis (atDjA30, atDjA2, atDjA54, atDjA24, atDjA26, and atDjA52, accessions BAB11067, NP_568412, NP_188410, NP_568076, NP_565533, and AAD55483, respectively), from yeast (ScYDJ1p and ScMDJ1p, accessions NP_014335 and NP_116638, respectively), and from the cyanobacteria Synechococcus sp. WH5701 (SynDnaJ, accession ZP_01084411) and Prochlorococcus marinus (PmDnaJ, accession YP_001013845). Phylogenetic analysis was conducted using version 4 of the MEGA program (Tamura et al., 2007) on the basis of alignments made by version 1.8 of the ClustalW program.