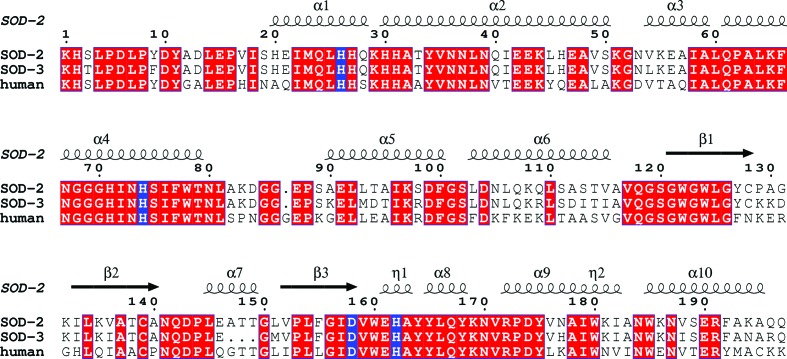
Figure 1.
Protein-sequence alignment of the three manganese superoxide dismutases MnSOD-2 (SOD-2), MnSOD-3 (SOD-3) and human MnSOD (Borgstahl et al., 1992 ▶) with the secondary-structure elements of MnSOD-2 superposed. The two domains extend between residues 1 and 81 (N-domain), forming an α-hairpin with a turn between α2 and α3, and residues 90 and 197 (C-domain). Helix α7 is absent in MnSOD-3 owing to the deletion of residues 147–149. The alignment figure was made using the program ESPript (Gouet et al., 1999 ▶). Red boxes indicate strictly conserved residues. The helices labelled η1 and η2 represent 310-helix segments. Residues that are ligands to the manganese are boxed in blue.