Table 1. Crystallographic summary of the structures of MnSOD-2 and MnSOD-3.
Values in parentheses are for the outermost shell.
| SOD-2 (100K) | SOD-3 (100K) | SOD-3 (293K) | |
|---|---|---|---|
| Resolution range () | 69.81.8 (1.91.8) | 44.11.7 (1.791.7) | 57.641.77 (1.871.77) |
| Space group | P41212 | P41212 | P41212 |
| Unit-cell parameters () | a = b = 81.0, c = 137.4 | a = b = 81.8, c = 136.0 | a = b = 81.5, c = 138.0 |
| No. of observed reflections | 172926 | 181809 | 190572 |
| No. of unique reflections | 41573 | 49346 | 45989 |
| Redundancy | 4.2 (4.2) | 3.7 (3.6) | 4.1 (4.0) |
| Completeness (%) | 97.2 (98.9) | 96.2 (95.8) | 99.9 (99.9) |
| I/(I) | 8.7 (5.1) | 5.0 (2.1) | 8.6 (2.3) |
| R merge † (%) | 5.6 (14.4) | 7.6 (36.4) | 6.2 (33.8) |
| Refinement and model statistics | |||
| Resolution range for refinement () | 43.991.8 | 44.101.7 | 70.191.77 |
| R factor (%) | 16.9‡ | 18.9 | 21.6 |
| R free § (%) | 20.1‡ | 22.6 | 26.2 |
| No. of protein non-H atoms | 3170 | 3138 | 3138 |
| No. of water molecules | 448 | 420 | 298 |
| No. of manganese ions | 2 | 2 | 2 |
| No. of sulfate atoms | 15 | 0 | 0 |
| No. of malonate-ion atoms | 0 | 7 | 0 |
| R.m.s.d. bond lengths¶ () | 0.009 | 0.013 | 0.015 |
| R.m.s.d. bond angles¶ () | 1.1 | 1.4 | 1.4 |
| B factors (2) | |||
| Overall | 16 | 19 | 23 |
| Protein | 14 | 18 | 22 |
| Water | 29 | 32 | 26 |
| Manganese | 14 | 15 | 15 |
| Sulfate | 33 | 0 | 0 |
| Malonate | 0 | 26 | 0 |
| Ramachandran analysis†† (%) | |||
| Residues in most favoured regions | 92.2 | 91.5 | 92.7 |
| Residues in additional allowed regions | 6.7 | 7.3 | 6.1 |
| Residues in generously allowed regions | 1.2 | 1.2 | 1.2 |
R
merge = 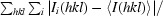
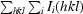 .
.
The R free set from the MnSOD-3 (100K) refinement was not transferred to the MnSOD-2 structure, leading to a slightly lower difference between R and R free, but they both fell normally during rebuilding of side chains that differ between the two homologous structures.
R free was calculated using 5% of the reflections that were set aside randomly.
Based on the ideal geometry values of Engh Huber (1991 ▶).
Ramachandran analysis using PROCHECK (Laskowski et al., 1993 ▶).
