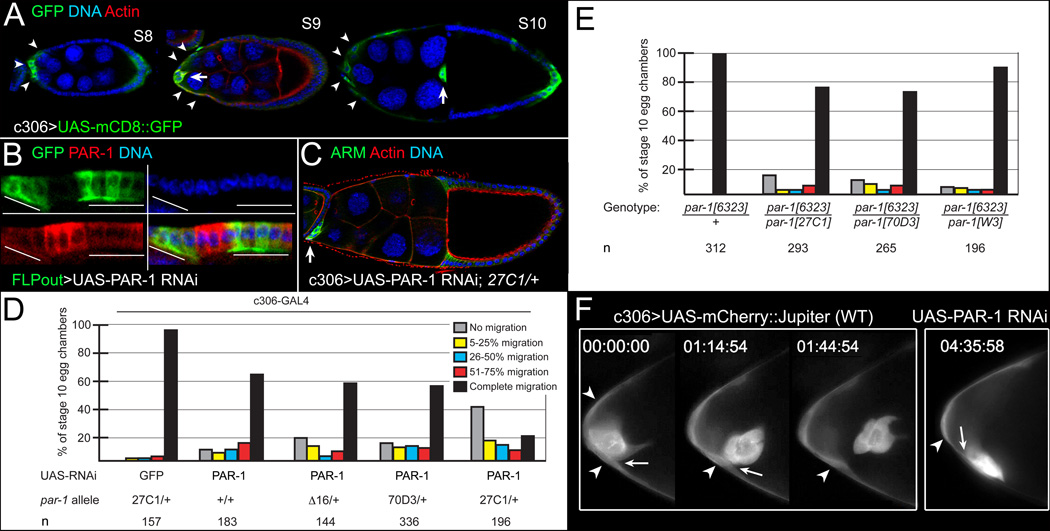
Figure 2. par-1 is required for border cell detachment.
(A–D) Downregulation of PAR-1 by UAS-PAR-1 RNAi disrupts border cell detachment. (A) Stage 8–10 egg chambers showing c306-GAL4 driver pattern, visualized by UAS-mCD8::GFP (green), in anterior stretched follicle cells (arrowheads) and border cells (arrows). (B) Follicle cells expressing UAS-PAR-1 RNAi in FLPout GAL4 clones, marked by GFP (green, lines), have reduced PAR-1 (red). (C) Stage 10 par-127C1/+ egg chamber expressing PAR-1 RNAi driven by c306-GAL4, stained for ARM (green) and F-actin (red) to label cell membranes and DAPI (blue) to label nuclei. The border cells (arrow) did not migrate. (D, E) Quantification of border cell migration defects in wild-type or par-1/+ egg chambers expressing UAS-PAR-1 RNAi driven by c306-GAL4 (D), and in viable par-1 mutant egg chambers (E). Percentage of border cells that did not migrate/detach (gray) or migrated 5–25% (yellow), 26–50% (blue), 51–75% (red), or 76–100% (black) of normal distance to the oocyte; n, number of egg chambers examined. (F) Frames from time-lapse movies of stage 9 control (Movie S1) or PAR-1 RNAi (Movie S2) egg chambers at the indicated time (h:min:s). Control border cells detached at ~1.5 hours; PAR-1 RNAi border cells (n = 10) stayed attached (arrows) to follicle cells (arrowheads).