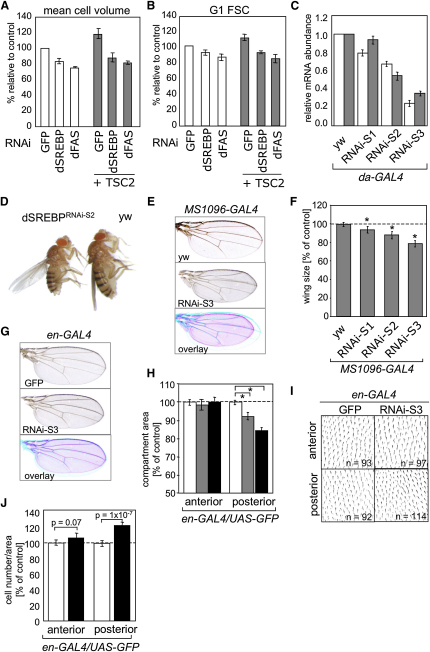
Figure 5.
Silencing of dSREBP Reduces Cell and Organ Size in Drosophila
(A) Kc167 cells were treated with dsRNAs corresponding to GFP, dSREBP, or dFAS in the absence or presence of dTSC2 dsRNA for 5 days, and electronic cell volume was determined.
(B) Cells treated as in (A) were used to determine FSC-A of the G1 population by FACS.
(C) dSREBP RNAi was expressed ubiquitously during embryogenesis using the daughterless-GAL4 (da-GAL4) driver. Expression of dSREBP (white bars) and dFAS (gray bars) in second instar larvae was determined by qPCR. Transcript levels are normalized to actin mRNA levels and represent three independent experiments. (RNAi-S1, single insertion, RNAi-S2 and RNAi-S3, double insertions).
(D) Representative phenotype of female flies expressing dSREBP RNAi using the da-GAL4 driver compared to control (yw).
(E) Wings of flies expressing dSREBPRNAi-S3 in the dorsal cell layer of the wing.
(F) Wing area analysis of control (yw), dSREBPRNAi-S1, dSREBPRNAi-S2, and dSREBPRNAi-S3. (∗) p ≤ 10−5.
(G) Wings of flies expressing dSREBPRNAi-S3 in the posterior compartment of the developing wing.
(H) Changes in wing area of control flies (white bars) or flies expressing SREBP RNAi-2 (gray bars) or SREBP RNAi-3 (black bars) in the posterior compartment of the wing. (∗) p ≤ 10−8.
(I) High-magnification images of the anterior and posterior compartments of wings shown in (G).
(J) Changes in wing epithelial cell number of flies expressing GFP (white bars) or dSREBPRNAi-S3 (black bars) in posterior compartment of the wing. Error bars represent SD.