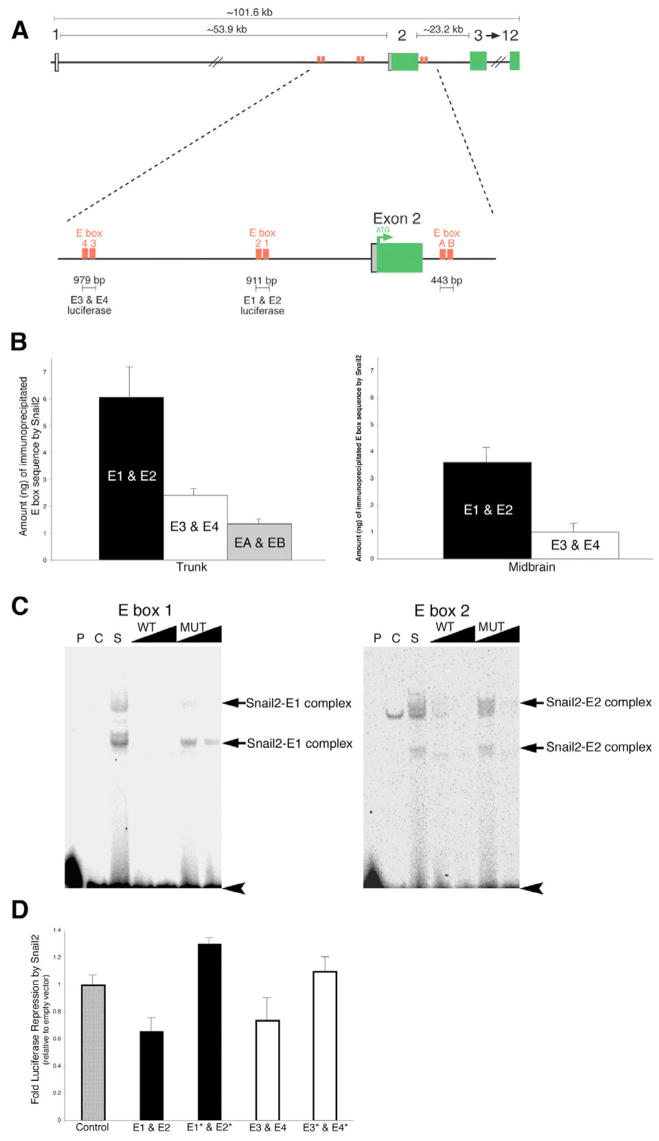
Fig. 4. Snail2 binds specifically to E boxes surrounding the ATG codon of Cad6B.
(A) Chick Cad6B is encoded by 12 exons on chromosome 2. Sequence analysis identified six E boxes with the motif CAGGTA clustered upstream and downstream of the ATG located in exon 2 of Cad6B. Primers and probes for chromatin immunoprecipitation (ChIP) and QPCR assays were designed to anneal to the region between E boxes within each clustered E-box pair. For luciferase experiments, the region of sequence spanning each clustered pair of E boxes that was cloned into the luciferase reporter vector is indicated. (B) ChIP experiments identify the preferential association of Snail2 with E boxes in the premigratory neural crest region of the chick trunk and midbrain, with E1 and E2>E3 and E4>EA and EB. At each axial level, the experiment was repeated at least three times, and the results presented are representative of a typical experiment. No association of Snail2 to distal sequences lacking E-box motifs in exon 11 of the Cad6B coding region was observed, and no E-box motifs were amplified by QPCR performed on samples immunoprecipitated with control antibodies (GFP, IgG) or with no antibodies added. (C) Electrophoretic mobility shift assay (EMSA) identifying the specific interaction of Snail2 with E box 1 and E box 2 in the Cad6B regulatory region (see also Fig. S3 in the supplementary material). Double-stranded 33P-end-labeled E-box oligonucleotide probes were incubated with control or Snail2 nuclear extract in the presence or absence of various competitor DNA oligonucleotide probes. P, no protein; C, 10 μg control nuclear extract. In the remaining lanes, 10 μg Snail2 nuclear extract was added to each binding reaction. S, no competitors added; WT and MUT, 10-fold and 100-fold molar excess of either unlabeled wild-type or unlabeled mutant E-box probes added, respectively. Retarded Snail2–E-box complexes are identified by arrows, and unbound probes are indicated by arrowheads. Two bands corresponding to E-box complexes were observed in the presence of the Snail2 nuclear extract. These bands were competed by the addition of wild-type E-box oligonucleotides, but to a much reduced efficiency with mutated E-box oligonucleotides. (D) Luciferase assays demonstrate that Snail2 represses luciferase expression from wild-type Cad6B E box-luciferase reporters but not from reporters with mutated E boxes. E1* and E2*, E1 and E2 double mutant; E3* and E4*, E3 and E4 double mutant. Results are reported as fold repression (relative to empty vector), and are an average of at least three independent QPCR experiments.