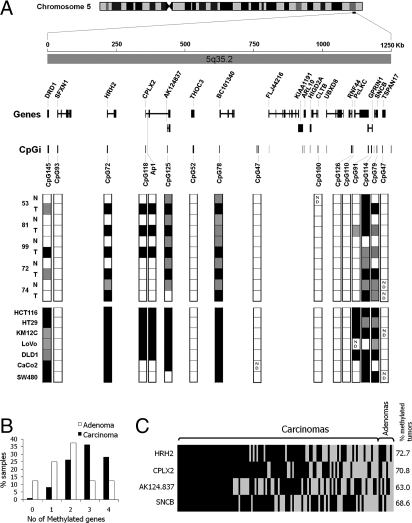
Fig. 1.
(A) Genomic map of the 5q35.2 region covered in this study. The region spanned 1.25 Mb and included 18 genes from the most centromere-proximal DRD1 to most telomere-proximal TSPAN17. CpG islands (CpGi) across 5q35.2 are named according to the number of CpG sites they contain, except for the Ap1 fragment, which is not a CpG island itself. Bisulfite sequencing data from the five normal-tumor pairs and the seven colorectal cancer cell lines is summarized. Each box represents a CpG island, which can be found unmethylated (white), partially methylated (gray), or heavily methylated (black). ND, not determined. The complete DNA methylation dataset is found in Fig. S2. (B) Methylation data summary for the 4 analyzed genes (HRH2, CPLX2, AK124.837, and SNCB) in 118 normal-tumor (110 carcinomas, 8 adenomas) pairs. (C) Individual data for carcinomas and adenomas sorted by methylation frequency. A white box indicates unmethylated status and a black box methylated status. The methylation status was ascertained by melting curve analysis and confirmed by bisulfite sequencing in a subgroup of 50 samples.