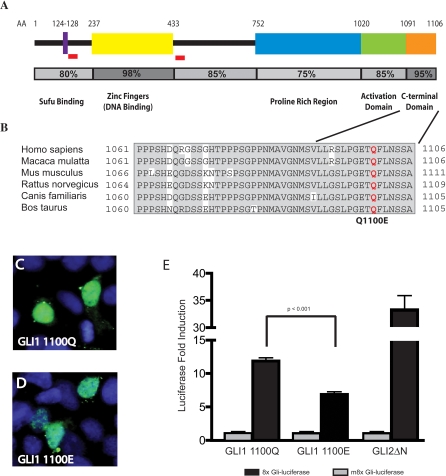
Figure 2. Q1100E Disrupts a Conserved Region of the GLI1 Protein and Reduces GLI1 Transcriptional Activity.
(A) Conservation of known functional domains in the Gli1 protein. Previously described Sufu binding, DNA binding, and transactivation domains [34,57,58] are shown schematically. Amino acid conservation of each domain is represented numerically and by shading of the bar below the domain. Red boxes indicate regions known to regulate GLI1 protein stability [49]. The conserved C-terminal domain that includes Q1100E is adjacent to a known transactivation domain.
(B) Alignment of the C terminus of mammalian Gli1 proteins. This region (AA 1080–1106) is highly conserved in mammalian lineages.
(C, D) GLI1 Q1100 and E1100 have similar cellular localization in 293T cells.
(E) GLI1 E1100 is deficient in driving activation of the 8xGli-Luciferase reporter compared to GLI1 Q1100. GLI2ΔN is a strong activator of 8xGli-Luciferase and serves as a positive control for GLI1 activation. The m8xGli-Luciferase construct contains eight mutant Gli binding sites and serves as a negative control. Data are shown from six triplicate experiments done using two different plasmid preparations (n = 18). Bar heights indicate mean fold activation above baseline, and error bars indicate standard error of the mean. p-Values were calculated using the Student's t-test.