Fig. 3.
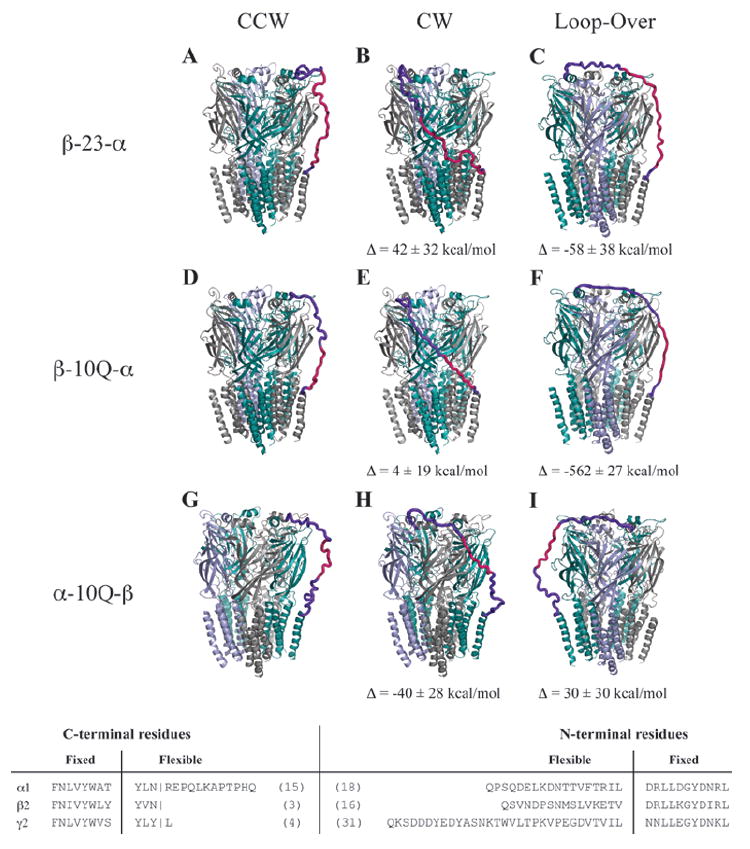
Hypothetical tandem linker conformations. Several linker architectures reported in the literature for GABAARs were evaluated for linker strain using molecular mechanics models. Here, we present the optimized tandem-containing receptors in each possible tandem configuration: counterclockwise (left), clockwise (center), and loop-over (right). The color code for subunits is α1 (teal), β2 (gray), and γ2 (blue). Linkers are hot pink, and regions of the native subunit allowed flexibility are shown in purple. Evaluated linkers were β-23-α (A–C) with linker composed of Q3(Q2A3PA)2AQ5, β-10Q-α (D–F), α-10Q-β (G–I), all tested in oocytes by Baumann and coworkers (9). Also tested but not shown were α-(Q9+signal peptide)-β (11,12), β-(10Q+signal peptide)-α from Fig. 2, and γ-26-β (20). The latter three showed no energetic differences in linker strain for counterclockwise, clockwise, or loop-over conformations. To see differences in relative strain among the tandem arrangements for each linker, ΔE was calculated by subtracting the converged potential energy of linker-imposed strain from the energy for the counterclockwise conformation. Data are mean ± SD from three or more converged cycles. For these calculations, all parts of the pentamer were held fixed except for linker sequence and adjoining terminal regions (3-31 residues, bottom). Linker and signal peptides were added to our core model and assigned Gasteiger-Marsili charges and Sybyl atom types from the Tripos forcefield by default. Each structure was minimized 4000 steps by default methods and was then subjected to 8 to 15 2-picosecond cycles of simulated annealing in vacuo with a linear temperature gradient from 700 to 200 K, followed by a quenching minimization after each cycle. Cycles were repeated until apparent convergence of system potential energy.
