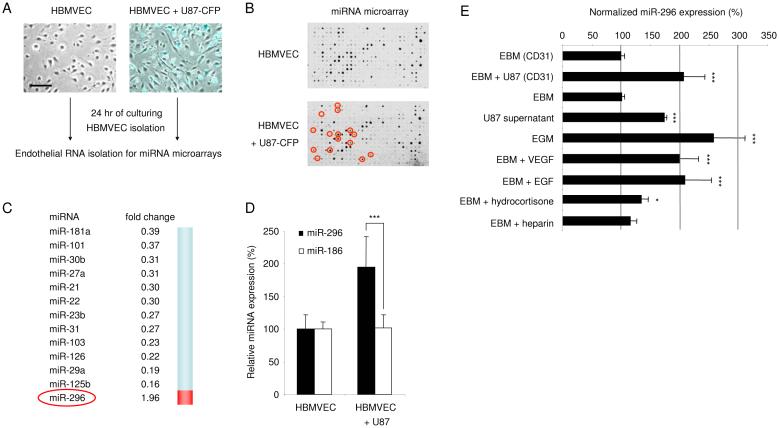
Fig. 1.
Glioma-induced disregulation of miRNAs in human brain endothelial cells (A) Primary human brain microvascular endothelial cells (HBMVEC) were cultured in the absence (left) or presence (right) of human U87-CFP glioma cells. Images were produced by using a combination of light and fluorescence microscopy, size bar 100 μm. (B) Array hybridization analysis of miRNAs extracted from CD-31+ cells, sorted from HBMVECs cultured without (upper array) or with (lower array) U87-CFP glioma cells for 24 hr. The density of the hybridization signals (black spots) reflects the relative expression level of particular miRNAs. Red circles indicate miRNAs which are significantly altered by co-culture with glioma cells. (C) A list of significantly decreased (fold change <0.5) or increased (fold change >1.9) miRNAs in HBMVECs exposed to U87-CFP glioma cells. (D) Overexpression of miR-296 was confirmed by qRT-PCR analysis. RNA extracted from CD31+ HBMVECs cultured in the absence or presence of U87 glioma cells was analyzed by qRT-PCR. The data were normalized to the level of GAPDH mRNA in each sample. (E) HBMVECs were cultured in the presence or absence of U87 glioma cells, isolated using CD31 beads, and subjected to miR-296 qRT-PCR. Alternatively, HBMVECs were cultured in various culture media and subjected to miR-296 qRT-PCR, the miR-296 levels were normalized to EBM (CD31). Error bars indicate S.D., *p < 0.05, ***p < 0.001, t test.