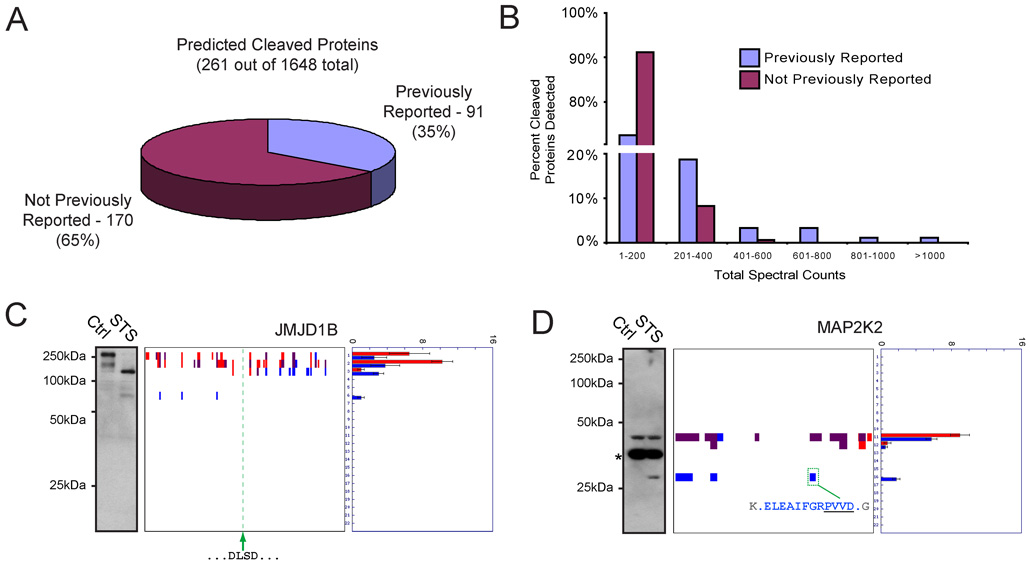
Figure 3. Global analysis of cleaved proteins in apoptotic cells.
(A) A total of 261 predicted cleaved proteins were identified by PROTOMAP in apoptotic cells, 91 (35%) of which corresponded to established caspase substrates and/or proteins known to be proteolyzed during apoptosis. The remaining 170 proteins (65%) were not previously known to be cleaved during apoptosis. (B) Comparison of the spectral count values for previously known versus unknown cleaved proteins in apoptotic cells. Note that proteins with high spectral count values were predominantly from the former group. (C) and (D) Examples of predicted cleaved proteins that were confirmed by western blotting. In the case of JMJD1B (C), an implicit caspase cleavage sequence at amino acids 817–823 (tandem DLSD) could be identified that resided in between the persistent 75 kDa N- and 100–125 kDa C-terminal fragments. In the case of MAP2K2 (D), the explicit caspase cleavage site was identified by PROTOMAP at residue D284. The asterisk in the MAP2K2 western blot likely corresponds to antibody cross-reactivity with a background protein, as no spectral counts were detected for MAP2K2 in the corresponding gel band. Spectral count data are represented as the mean +/− SEM for four independent experiments.