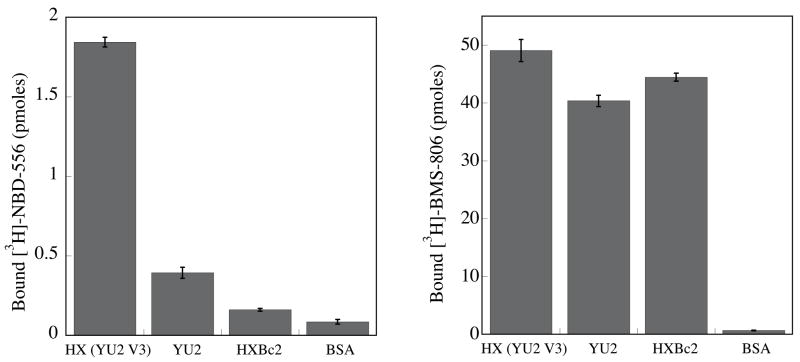
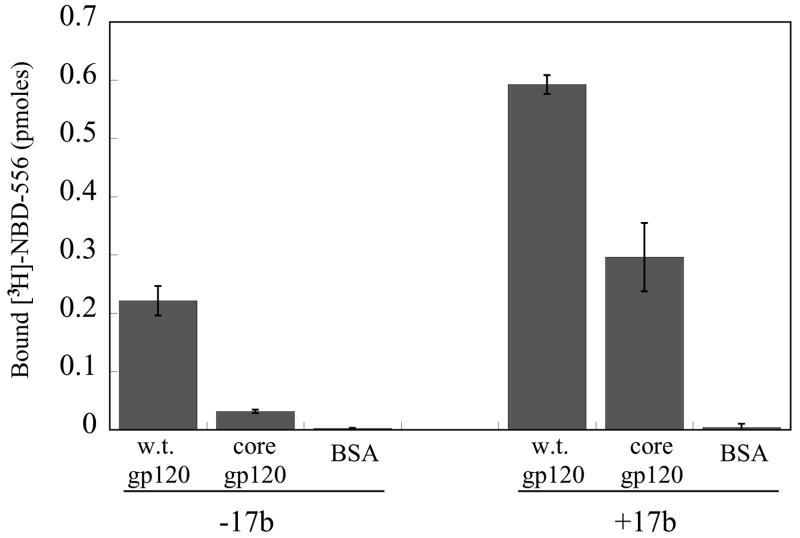
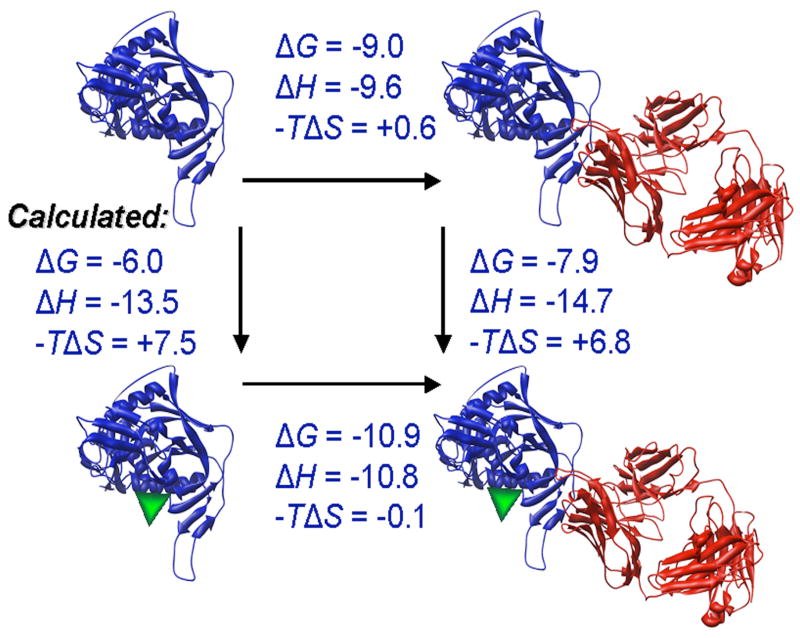
Figure 2. Binding of NBD-556 to the conserved core of HIV-1 gp120.
a. The binding of [3H]-NBD-556 to the indicated HIV-1 gp120 envelope glycoprotein variants or to bovine serum albumin (BSA) is shown (left panel). The right panel shows binding of the same gp120 envelope glycoproteins to [3H]-BMS-806 as a control. The values shown are the means+/−SEM from one experiment (n=3). b. The binding of [3H]-NBD-556 to the HIV-1YU2 w.t. gp120 glycoprotein or to the gp120 core protein in the absence or presence of the 17b antibody (1 mg/ml) is shown. Binding of [3H]-NBD-556 to the BSA control protein is shown for comparison. The values shown are the means +/− SEM from a single experiment (n=3). The experiment was performed three times with comparable results. c. The thermodynamic cycle for the binding of NBD-556 and the 17b antibody to the HIV-1YU2 gp120 core was studied by titrating the gp120 core with 17b in the presence of a saturating concentration of NBD-556. The values associated with the direct binding of NBD-556 to the gp120 core were calculated by completing the thermodynamic cycle. The structures of the gp120 core and the 17b Fab fragment (PDB entry 1GC1) are depicted in blue and red, respectively. NBD-556 is represented by the green triangle.