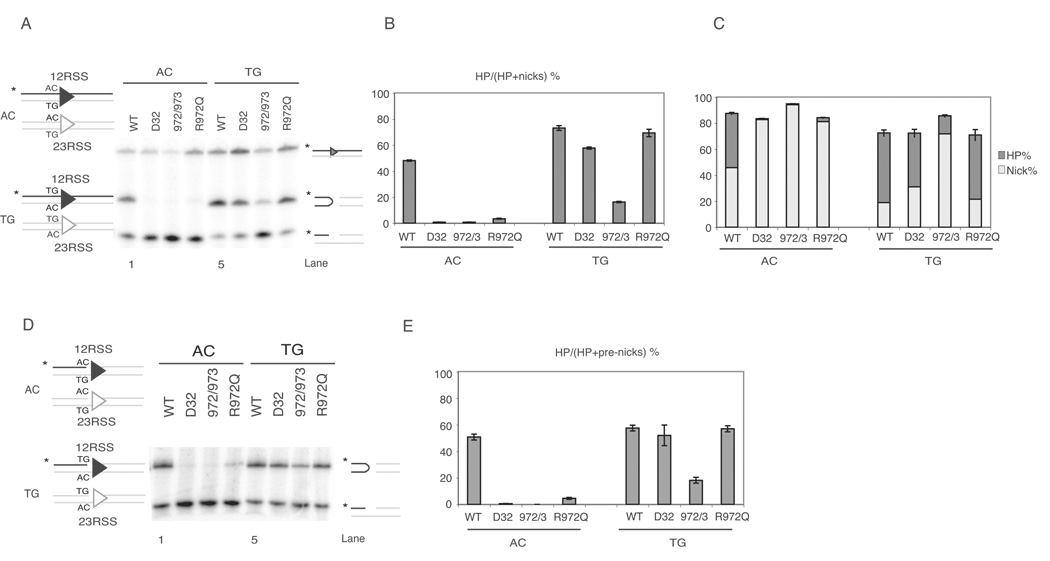
Figure 1. R972Q RAG1 mutant shows preferences for coding flank sequence in vitro.
(A) Cleavage assay using oligonucleotide substrates as depicted on the left, with either AC (non-preferred) or TG (preferred) flank. Only one strand of each oligo pair was 5’-radiolabled (*, strand in black), and thus detectable after electrophoresis.
(B) Quantitative analysis of gel in (A), showing the percentage of hairpin formation. Error bars represent standard error of the mean of three experiments.
(C) Quantitative analysis of gel in (A). The percentage of hairpins and nicks are shown in dark grey and light grey, respectively, and the length of each bar represents total nicking activity, as nicks were converted into hairpins.
(D) Cleavage assay using oligonucleotide substrates with same sequence as in (A), except these contain a preformed nick.
(E) Quantitative analysis of gel in (D), showing the percentage of pre-nicks converted to hairpins.