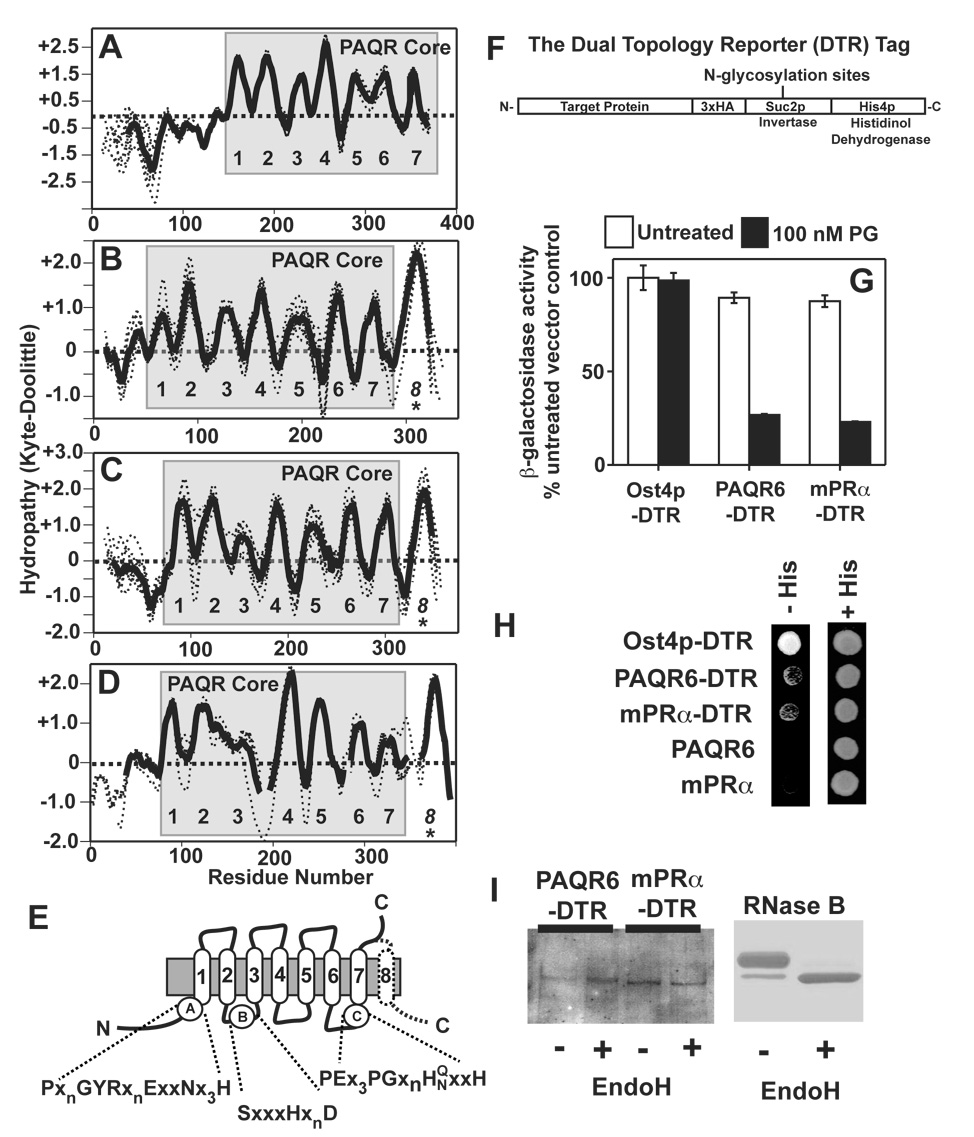
Figure 5. Topological analysis of Class I and Class II PAQRs. (A–D).
Hydropathy plots for the individual proteins analyzed in Figure 3 were generated and aligned (dotted lines). An average hydropathy plot for all members of each clade were generated (solid lines). The core PAQR motif is shaded in grey and the predicted TMs are numbered. All vertebrate members of the AdipoR1 and AdipoR2 clade (A), the mPRγ/PAQR6 clade (B), the mPRα/mPRβ clade (C) and the PAQR9 homologs (D). (E) Predicted topology of the core PAQR motif with the locations of the three highly conserved motifs shaded in black in Figure 4. The predicted eighth TM in the mPRs is shown with a dotted line. (F) The structure of the dual topology reporter tag. (G) DTR-tagged PAQR6 and mPRα expressed in the pJK90 plasmid repress FET3-lacZ in response to 100 nM progesterone in cells grown in medium containing 0.05% galactose/1.95% raffinose. (H) Rescue of the histidine auxotrophy of the STY50 strain by DTR-tagged PAQR6 and mPRα on plates containing histidinol. Plasmids expressing untagged versions of PAQR6 and mPRa are shown as negative controls. A plasmid expressing DTR-tagged Ost4p is shown as a positive control. (I) Membrane extracts from cells expressing DTR-tagged PAQR6 or mPRα are either left untreated (−) or treated with the endoglycosidase EndoH (+) and subsequently run on protein gels and transferred to nitrocellulose membranes for Western blot. anti-HA antibodies were used to detect the DTR tag. (left) Coomassie stained gel of EndoH treated RNase B under the same conditions as those used to treat the cell membrane extracts is shown on the right as a positive control.