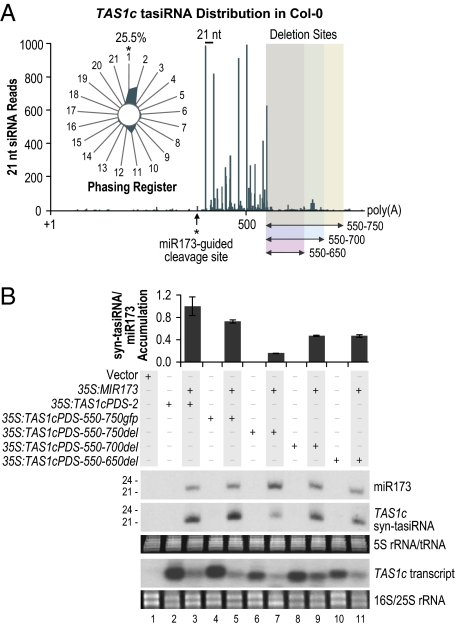
Fig. 5.
The effect of internal deletions on syn-tasiRNA formation. (A) tasiRNA distribution across the endogenous TAS1c transcript in Col-0. 5,888 TAS1c tasiRNA reads from Col-0 leaf and whole plant Illumina 1G sequencing data sets were obtained from the Arabidopsis Small RNA Project database (31). (Inset) The radar plot displays the percentages of reads corresponding to each of the 21 registers, with the 5′ end formed by miR173-guided cleavage defined as register 1. Internal deletion positions are shown by the shaded boxes. (B) Blot assays. Parental 35S:TAS1cPDS-2 and modified constructs containing internal deletions were expressed individually or in combination with 35S:MIR173 as indicated above the blot panels. Mean relative level +/− SEM of syn-tasiRNA relative to miR173 (35S:TAS1cPDS-2 + 35S:MIR173 = 1.0). One of three biological replicates is shown. EtBr-stained 5S rRNA/tRNA and 18S/25S rRNA are shown as loading controls.