Figure 1.
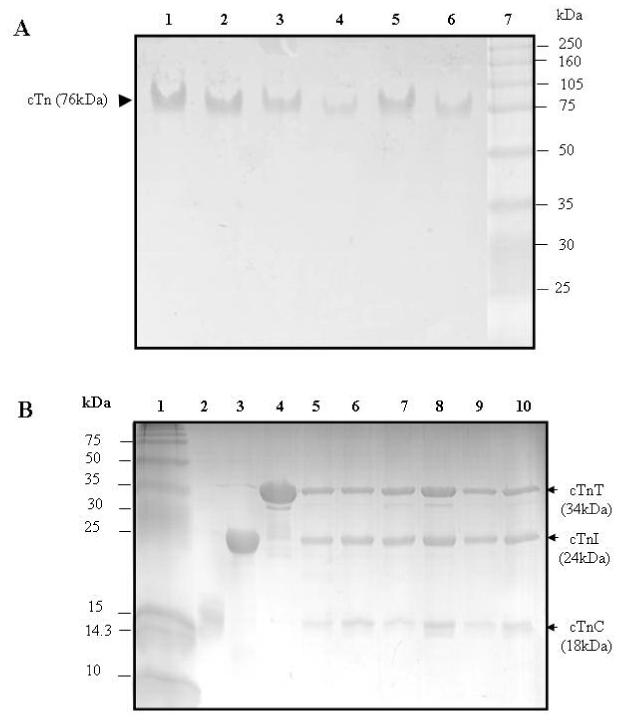
Electrophoresis analysis of reconstituted cardiac troponin
Panel A: Native PAGE gels (8% resolving and 4% stacking). Lanes 1-6, troponin reconstituted from wild-type cTnC, wild-type cTnT, and a cTnI mutant in which the single-cysteine was labeled with AEDANS. Lane 7, HMW protein standards (Amersham Biosciences). (1) cTnI(131C), (2) cTnI(151C), (3) cTnI(160C), (4) cTnI(167C), (5) cTnI(188C), and (6) cTnI(210C). These gels showed all six reconstituted samples ran as a single component with a mass of ∼ 76 kDa.
Panel B: SDS-PAGE gels (18% resolving and 4% stacking). Lane 1, LMW protein standards (Amersham Bisciences). (2) wild-type cTnC, (3) wild-type cTnI, and (4) wild-type cTnT. Lanes 5-10, troponin reconstituted from wild-type cTnC, wild-type cTnT, and a cTnI mutant in which its single cysteine was labeled with AEDANS. (5) cTnI(131C), (6) cTnI(151C), (7) cTnI(160C), (8) cTnI(167C), (9) cTnI(188C), and (10) cTnI(210C). The six reconstituted troponin samples were each resolved into three bands. Densitometry analysis of the bands was done with Bio-Rad Quantity One Software, and the results showed that the three resolved bands corresponded to cTnC, cTnI, and cTnT with a molar ration of 1:1:1. No significant degradation products were found on the gels.
