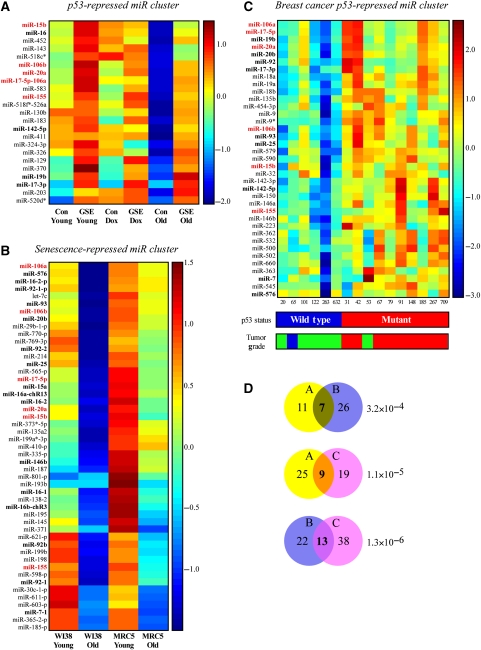
Figure 1.
miRNA clusters derived from three different datasets. The figure depicts three different expression matrices for miRNA clusters that originated from three microarray experiments. miRs appearing in all three clusters are indicated in red bold font. miRs appearing in two clusters are indicated in black bold font. (A) The ‘p53-repressed miR cluster.' Primary WI-38 cells that were infected with the p53-inactivating peptide GSE56 (GSE) and their empty vector control counterparts (Con) were analyzed for miRNA expression at early passage (Young), after doxorubicin treatment (0.2 μg/ml, 24 h) of early passage cells (Dox), and at the onset of replicative senescence (Old). A cluster of miRs that were repressed by p53 at normal conditions and in senescent cells is presented. (B) The ‘Senescence-repressed miR cluster.' Primary WI-38 and MRC5 cells were analyzed for miRNA expression at early passage (Young) and at the onset of replicative senescence (Old). A cluster of miRs that were repressed upon senescence in both cell types is presented. (C) The ‘breast cancer p53-repressed miR cluster.' Human primary breast cancers described by Sorlie et al (2006) and Naume et al (2007) were analyzed for their miRNA profiles. A cluster of miRs, the expression of which was anticorrelated with the presence of a wild-type p53 in the tumor is presented. p53 status was determined using TTGE and sequencing of exons 2–11. Grading was performed using histopathological evaluation according to the modified Scarff–Bloom–Richardson method and is represented by blue for grade 1, green for grade 2 and red for grade 3. (D) Venn diagrams depicting the overlaps between cluster pairs. The values in each circle represent the number of miRs from the indicated cluster that was detected by the array corresponding to the second cluster. The values in the circle overlapping regions represent the number of miRs that are shared between the two clusters. Hypergeometric P-values on the size of the overlaps are provided.