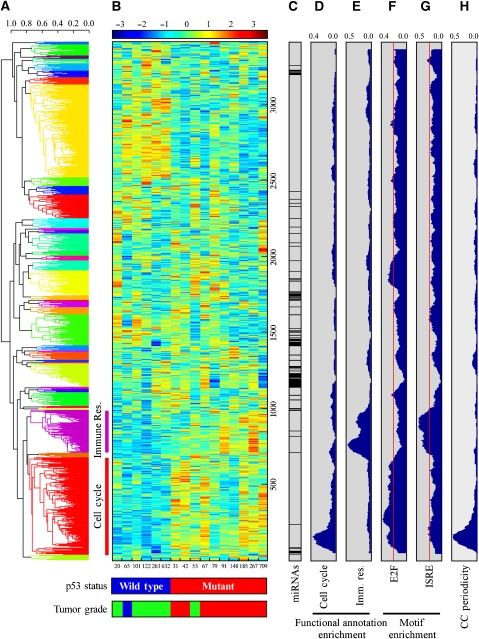
Figure 3.
Co-clustering of miRNA and mRNA expression data from human breast cancers. (A) A dendrogram for the expression data based on hierarchical clustering and average linkage. Data were clustered into 40 clusters, which are indicated by different colors of the dendrogram. miRs from the ‘breast cancer p53-repressed miR cluster' were mapped to the red (cell cycle) and purple (immune response) clusters. (B) Expression matrix of the mRNAs and miRNAs analyzed. For p53 status and tumor grade analyses, see Figure 1 legend. Breast cancer samples are indicated by numbers below the matrix. (C) The bar indicates the position of miRs along the expression matrix. (D, E) Functional annotation analysis for ‘cell cycle' (C) and ‘immune response' (Imm. Res.) (D) terms. The plots represent the density (from 0 to 1) of mRNAs corresponding to each annotation term in windows of 100 genes. (F, G) Density plots for the appearance of the E2F and ISRE motifs, the most enriched elements in the cell cycle and immune response co-clusters, respectively. Red lines indicate the background levels of each motif, calculated as the fraction of genes in the genome containing the motif. (H) Density plot for cell-cycle periodic genes as defined by Whitfield et al (2002).