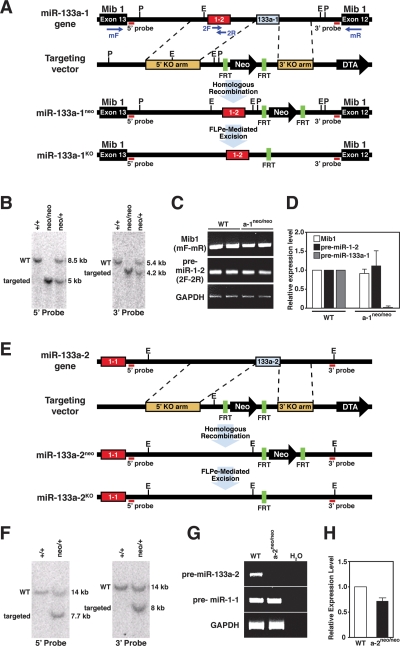
Figure 2.
Generation of miR-133a-1 and miR-133a-2 mutant mice. (A) Strategy for targeting of miR-133a-1. The miR-1-2/miR-133a-1 gene is located within intron 12 of the mind bomb 1 gene (Mib1). The genomic structure, targeting vector, and targeted allele for miR-133a-1 are shown. Pre-miR-133a-1 (68 bp) was replaced with a neomycin resistance cassette flanked by FRT sites, which allowed for FLPe recombinase-mediated excision. Probes for Southern blot analysis and primer positions for RT–PCR are shown. (E) EcoRI; (P) PstI. (B) Southern blot analysis for wild-type (WT) and miR-133a-1 mutant mice. Genomic tail DNA from agouti offspring was digested with PstI and probed with the indicated 5′ probe. The wild-type band migrated at 8.5 kb, and the targeted mutant band migrated at 5 kb, indicative of proper transmission of the targeted allele. Tail DNA was also digested with EcoRI and probed with the indicated 3′ probe. The wild-type band migrated at 5.4 kb, and the targeted mutant band migrated at 4.2 kb, confirming germline transmission of the targeted allele. Genotypes of mice are shown on top. (C) Expression of Mib1 and pre-miR-1-2 in wild-type and miR-133a-1 mutant mice detected by RT–PCR. RNA was isolated from hearts of adult wild-type and miR-133a-1 mutant mice (n = 2 for each genotype). Primer positions from A are shown in parentheses at the left. Primers 2F and 2R for miR-1-2 were located within the pre-miR-1-2 sequences. GAPDH mRNA was detected as a loading control. (D) Expression of Mib1, pre-miR-1-2, and pre-miR-133a-1 in wild-type and miR-133a-1 mutant mice detected by real-time PCR. Expression levels of each gene in miR-133a-1 mutant mice were normalized to GAPDH before comparison with expression in wild-type mice (n = 3 for each genotype). The Y-axis represents relative expression level compared with wild-type mice. Error bars indicate SEM. Pre-miR-133a-1 is not detected in miR-133a-1 mutant mice. (E) Strategy for targeting of miR-133a-2. The genomic structure, targeting vector, and targeted allele for miR-133a-2 are shown. Pre-miR-133a-2 (108 bp) was replaced with the neomycin resistance cassette flanked by FRT sites, which allowed for FLPe recombinase-mediated excision. Probes for Southern blot analysis are shown. (E) EcoRI. (F) Southern blot analysis for wild-type and miR-133a-2 mutant mice. Tail DNA from agouti off spring was digested with EcoRI and probed with the indicated 5′ and 3′ probes. Wild-type bands migrated at 14 kb and targeted mutant bands migrated at 7.7 kb using the 5′ probe and 8 kb using the 3′ probe. Genotypes of mice are shown on top. (G) Expression of pre-miR-133a-2 and pre-miR-1-1 in adult wild-type and miR-133a-2 mutant mice detected by RT–PCR. Primers for pre-miR-133a-2 and pre-miR-1-1 were located within their pre-stem–loop sequences. GAPDH levels were detected as a loading control. (H) Expression levels of pre-miR-1-1 in wild-type and miR-133a-2 mutant mice detected by real-time PCR. Expression of pre-miR-1-1 in miR-133a-2 mutant mice was normalized to GAPDH before comparison with expression in wild-type mice (n = 3 for each genotype). The Y-axis represents relative expression levels compared with wild-type mice. Error bars indicate the SEM.