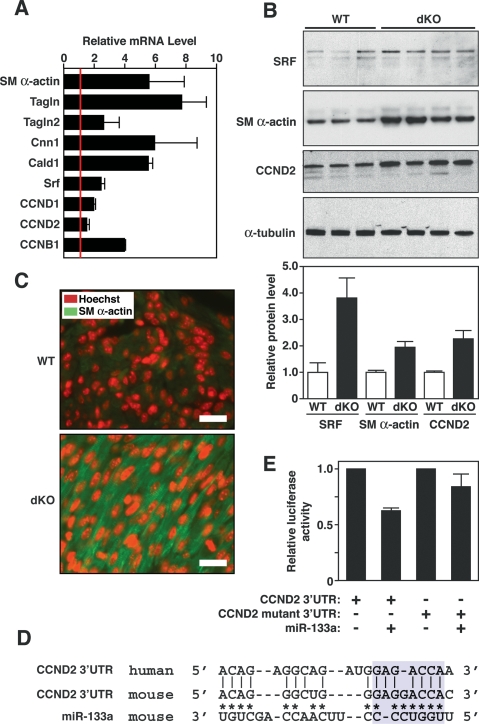
Figure 6.
Modulation of miR-133a targets in dKO hearts. (A) Expression of smooth muscle-specific genes and cyclin genes in hearts of wild-type and dKO mutant mice at P1 as detected by real-time PCR. Expression levels for each gene in dKO hearts were normalized to GAPDH and compared with wild-type hearts. Error bars indicate the SEM. (SM α-actin) Smooth muscle α-actin; (TAGLN) transgelin (also called SM22); (TAGLN2) transgelin 2 (also named SM22β); (CNN1) calponin I; (CALD1) caldesmon; (CCND1) cyclin D1; (CCND2) cyclin D2; (CCNB1) cyclin B1. (B) Expression of SRF, SM α-actin, and CCND2 in wild-type and dKO mutant hearts. Western blot analysis was performed on hearts from P1 wild-type (n = 3) and dKO mutant (n = 4) mice. α-Tubulin was detected as a loading control. Quantification of bands by densitometry showed a 3.5-fold and twofold increase in expression of SRF, SM α-actin, and CCN2 in dKO compared with wild-type hearts. (C) Increased SM α-actin expression in dKO hearts at P1. Histological sections of wild-type and dKO mutant hearts at P1 were stained for SM α-actin (green) and for nuclei with Hoechst (red). Pictures of wild-type and dKO hearts were taken under the same exposure parameters. Bar, 20 μm. Hoechst staining was reproducibly more intense in sections of dKO hearts compared with wild type, which may reflect greater DNA synthesis in the mutant. (D) Sequence alignment of the human and mouse cyclin D2 3′ UTR and miR-133a. Asterisks point to Watson-Crick base-pairing between mouse cyclin D2 3′ UTR and miR-133a. Base-pairing between miR-133a seed sequences with cyclin D2 3′ UTR is highlighted in blue. Mutations in cyclin D2 3′ UTR were introduced to disrupt base-pairing with the seed sequences. (E) Luciferase assay of cyclin D2 3′ UTR in Cos-1 cells. Wild-type and mutant cyclin D2 3′ UTR sequences were cloned into luciferase-reporter constructs and were cotransfected with a plasmid expressing miR-133a into Cos-1 cells. Forty-eight hours post-transfection, luciferase activity was measured and normalized to β-galactosidase activity. Error bars represent the SEM.