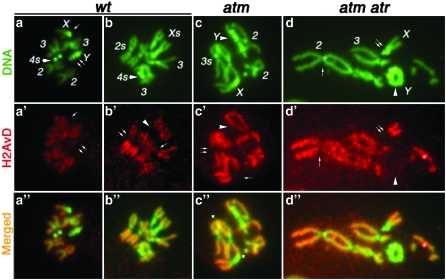
Figure 3.—
H2AvD distribution on mitotic chromosomes DNA is in green, with major chromosomes labeled (a–d). Anti-GFP signals are in red (a′–d′). Regions in which H2AvD-GFP is underrepresented are selectively labeled as follows: In the male wt nucleus (a, a′, and a″), anti-GFP did not stain X centric heterochromatin (single arrow) or the entire Y chromosome (double arrow). In the female wild-type (wt) nucleus (b, b′, and b″), H2AvD is missing for centric heterochromatin on chromosomes X (arrowhead in b′), 2 (double arrow), and 3 (single arrow). In the male atm nucleus (c, c′ and c″), H2AvD is missing on the Y chromosome (arrowhead), on X centric heterochromatin (single arrow), and on chromosome 3 (double arrow). In the male atm atr nucleus (d, d′, and d″), H2AvD is missing from Y (arrowhead), X centric heterochromatin (double arrow), and chromosome 2 centric heterochromatin (single arrow). In c″, several sites of telomere fusion are marked with an asterisk. In d″, several telomere fusions are present. However, the precise fusion points are difficult to pinpoint due to severe aneuploidy and gross genome rearrangement in atm atr nuclei. Nevertheless, the Y chromosome formed a ring due to telomere fusions.