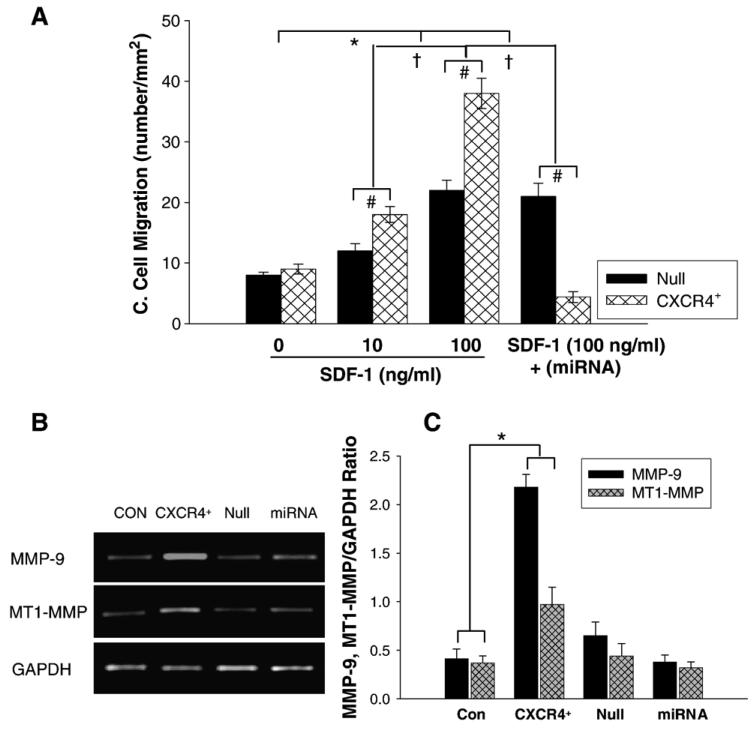
Fig. 3.
Cell migration and MMPs expression. A: Chemotaxis experiments were carried out by using transwells (5-μm pore). Cultured MSCs transduced with AdCXCR4/GFP (1 × 105) or Ad-miRNA targeting CXCR4 in 100 μl medium were added to the upper chamber, and 600 μl medium with or without SDF-1α (0, 10, 100 ng/ml) was placed in the bottom chamber. After 4 h incubation at 37 °C, migrating (bottom chamber) cells were counted. Exposure of Ad-CXCR4-MSCs to SDF-1α for 4 h resulted in strong cell migration, in a concentration dependent manner. The cell migration was significantly higher in Ad-CXCR4 group in the presence of SDF-1α (10 and 100 ng/ml) than in Ad-null group, p<0.05). The treatment of miRNA targeting CXCR4 prevented cell migration in response to SDF-1α, suggesting that CXCR4+-MSCs was functional and responded to SDF-1α. All values were expressed as mean±S.E.M. n=6 for each group. *p<0.05 vs. SDF-1α (0 ng/ml) group; †p<0.01 vs. other groups; #p<0.05 vs. null group. B: RT-PCR shows MMP-9 and MT1-MMP mRNA expression in CXCR4 overexpressing MSCs. CON indicates control MSCs group (lane 1); CXCR4+, CXCR4 overexpressing MSC group (lane 2); Null, adenoviral null treated MSCs (lane 3); miRNA, miRNA targeting CXCR4 (lane 4). GAPDH was used as the mRNA internal control to ensure equivalent loading. C: Quantitative assessment of CXCR4+-MSCs mRNA expression. All values were expressed as mean±S.E.M. n=6 for each group. *p<0.05 vs. control group.