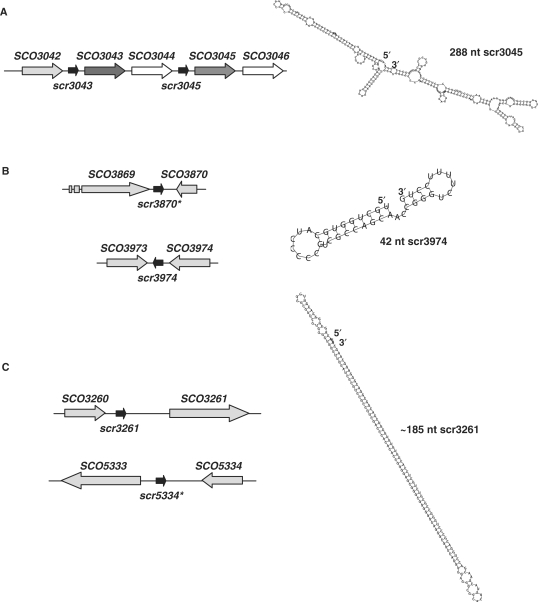
Figure 5.
Predicted structure and location of duplicated sRNA sequences within the S. coelicolor chromosome. (A) The sequence encompassing scr3045, SCO3045 and SCO3046 is extremely similar to that of scr3043, SCO3043 and SCO3044, suggesting a possible duplication event in S. coelicolor (other Streptomyces species have only one copy of each gene). Homologous genes are indicated in the same colours (scr3043 and scr3045: black arrows; SCO3043 and SCO3045: dark grey arrows; SCO3044 and SCO3046: white arrows). To the right is the predicted structure of scr3045. (B) The scr3974 sequence is extremely similar to that of a region found between SCO3869 and SCO3870, designated scr3870, although the flanking genes do not bear similarity to each other. To the right is the predicted structure of scr3974. (C) The scr3261 sequence bears striking similarity to a sequence located between SCO5333 and SCO5334, although, again, the flanking sequences are not similar. The scr3261 structure is distinctive due to its extensive internal complementarity. The asterisks in (B) and (C) indicate putative sRNA genes for which transcripts have never been experimentally detected.