Abstract
LldR (CGL2915) from Corynebacterium glutamicum is a transcription factor belonging to the GntR family, which is typically involved in the regulation of oxidized substrates associated with amino acid metabolism. In the present study, the crystal structure of LldR was determined at 2.05-Å resolution. The structure consists of N- and C-domains similar to those of FadR, but with distinct domain orientations. LldR and FadR dimers achieve similar structures by domain swapping, which was first observed in dimeric assembly of transcription factors. A structural feature of Zn2+ binding in the regulatory domain was also observed, as a difference from the FadR subfamily. DNA microarray and DNase I footprint analyses suggested that LldR acts as a repressor regulating cgl2917-lldD and cgl1934-fruK-ptsF operons, which are indispensable for l-lactate and fructose/sucrose utilization, respectively. Furthermore, the stoichiometries and affinities of LldR and DNAs were determined by isothermal titration calorimetry measurements. The transcriptional start site and repression of LldR on the cgl2917-lldD operon were analysed by primer extension assay. Mutation experiments showed that residues Lys4, Arg32, Arg42 and Gly63 are crucial for DNA binding. The location of the putative ligand binding cavity and the regulatory mechanism of LldR on its affinity for DNA were proposed.
INTRODUCTION
Corynebacterium glutamicum has been widely used for production of not only various types of amino acid, including l-glutamic acid and l-lysine, but also other substances, such as nucleotides and vitamins (1,2). To increase the industrial yield, many strategies have often been based on overcoming feedback regulation in the key step of the biosynthetic pathway in C. glutamicum (3–5). Further improvements require more systematic knowledge, including information regarding the factors influencing carbon flux and how to supply sufficient amounts of the necessary carbon precursors and coenzymes for specific synthesis of the target product. The release of the genomic sequences of C. glutamicum (accession numbers: AX114121, BA000036, BX927147) meets the need for such knowledge by facilitating extensive genome-wide analyses of amino acid biosynthesis (6–8).
Corynebacterium glutamicum can grow on various carbon sources (5,9). Glucose is the preferred carbon source, but it has been shown to be co-metabolized with other substrates, including l-lactate (10), acetate (11), pyruvate (12,13) and fructose (5,14). Nonhexose substrates, such as l-lactate, are taken up by the permease in a single step (10). However, sugar is taken up and simultaneously phosphorylated by the phosphoenolpyruvate (PEP)-dependent sugar phosphotransferase system (PTS) consisting of EI (encoded by ptsI)/HPr (ptsH) and a number of sugar-specific transmembrane permeases, i.e. EIIGlc/EIIMan (ptsG), EIIFru(ptsF) and EIISuc (ptsS), used for uptake of glucose/mannose, fructose and sucrose, respectively (6,15–18). Sugar phosphate formed and taken up through a PTS is the first intermediate in its metabolism. The transcriptional regulation of PTS proteins is particularly important because of their involvement in the first step of sugar metabolism and their central roles in selectively obtaining nutrition (18). Especially for C. glutamicum, the selection of carbon source has an important effect on not only its utilization but also on the metabolic pathway, subsequently influencing both yield and rate of amino acid production (9,19,20).
As one of the main raw materials for culturing C. glutamicum, industrial molasses usually contains 10–20% of its carbon source in the form of fructose (6). Moreover, uptake and concomitant phosphorylation of sucrose via EIISuc would yield glucose-6-phosphate (G-6-P) and fructose, which may be secreted into the medium. PTSfru (fructose-specific PTS) and PTSman (mannose-specific PTS) enable uptake and concomitant phosphorylation of fructose (6,21), and PTSfru is dominant. Uptake of fructose strongly enhances the formation of lactate excreted into the medium, which is a waste of a carbon source unless it can be reutilized (6). Stansen et al. (10) characterized an operon, cgl2917-lldD, indispensable for the utilization or re-utilization of l-lactate. cgl2917 (or NCgl2816) encodes a permease CGL2917 and lldD (cgl2918/NCgl2817) encode a quinone-dependent l-lactate dehydrogenase (LldD). During cultivation of C. glutamicum 2262 in continuous mode, excreted l-lactate is re-utilized after an increase in activity of LldD; such re-utilization may contribute to glutamate formation (22). Information regarding the regulation of fructose and l-lactate reutilization would be useful in a potential strategy for highly efficient amino acid production.
This report describes the structural and functional characterization of the CGL2915 protein (LldR) from C. glutamicum. This protein is a transcriptional repressor that regulates the expression of cgl2917-lldD and cgl1934-fruK-ptsF operons. In the latter operon, the genes encode the following deduced proteins: CGL1934, a putative transcriptional regulator of sugar metabolism; FruK (F-1-P kinase); and EIIfru. Of these proteins, CGL2917 and LldD are involved in the utilization of l-lactate and FruK and EIIfru in the utilization of fructose and sucrose. Intriguingly, the structure of LldR shows similarity to that of FadR (a fatty acid-responsive transcription factor) in dimer assembly by domain swapping; this is the first report of domain swapping in a dimeric transcription factor.
MATERIALS AND METHODS
Construction of recombinant LldR and mutant proteins
The gene lldR (NCgl2814) encoding LldR was amplified by PCR from genomic DNA of C. glutamicum ATCC 1332 using primer pairs corresponding to the predicted 5′ and 3′ termini. The PCR products were digested with NdeI and SalI, and then cloned into the pET26b vector (Novagen, San Diego, CA, USA), which had been digested with NdeI and XhoI. The resulting construct encoded LldR with a His6 tag (LEHHHHHH) at the C-terminus.
All expression vectors for mutant proteins were prepared with a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) using synthesized primers and the LldR expression vector described above as a template. DNA sequences were confirmed using an ABI PRISM 310 genetic analyser (Applied Biosystems, Tokyo, Japan).
Preparation of LldR and mutant proteins
The constructed expression vector was transformed into Escherichia coli strain B834 (DE3, methionine auxotroph). Transformants were then grown at 37°C in LB medium supplemented with 25 µg ml–1 kanamycin. The protein was expressed by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM at an OD600 of ∼0.6. After induction for 20 h at 25°C, the cells were harvested, re-suspended in buffer A (50 mM Na-Pi, pH 8.0, 0.5 M NaCl) and disrupted with a French press. The resulting suspension was centrifuged at 40 000 g at 4°C for 30 min to remove the debris. The supernatant was filtered through a 0.45-μm filter (Sterivex™-HV; Millipore, Billerica, MA, USA), then applied to a HiTrap chelating HP column (Amersham Biosciences Inc., Piscataway, NJ, USA) and eluted with a linear gradient of 0.0–0.5 M imidazole in buffer A. The fractions containing the target protein were sequentially purified by chromatography on a HiLoad 26/60 Superdex 200 pg column (Amersham Biosciences) with buffer A as the eluent. The target peak, containing LldR or mutant protein, was recovered and dialysed overnight against buffer B (20 mM Tris–HCl, pH 7.5, 0.2 M NaCl, 10% glycerol, 1 mM dithiothreitol), then concentrated and used for further experiments. For production of selenomethionine (Se-Met)-substituted LldR, cells were cultured in minimal medium containing Se-Met. The procedure for purification of Se-Met LldR was the same as that for the native and mutant proteins.
Crystallization and data collection
Crystal screening was performed in sitting drop mode by mixing 1 μl of protein (5 mg ml–1 in buffer B) with 1 μl of reservoir solution and equilibrating against 100 μl of reservoir solution at 20°C. Initial crystals were obtained with a CSI&II screening kit (No.22; Hampton Research, Aliso Viejo, CA, USA), and further optimization of conditions was performed using the hanging drop vapour diffusion method. Crystals grew to average dimensions of 0.2 × 0.6 × 0.1 mm within 2 weeks in 30% PEG 4000, 0.1 M Tris–HCl, pH 8.8, 0.1 M NaOAc.
Crystals were soaked in mother liquor containing an additional 15% glycerol for 30 s and flash-cooled under a stream of liquid nitrogen. Two diffraction data sets (Se-MAD and native) were collected below 100 K at BL41XU of SPring-8 (Hyogo, Japan) and BL6A of PF (Tsukuba, Japan), respectively. Crystals of Se-Met and native LldR belonged to space group C2221, with cell dimensions of a = 96.0, b = 107.3, c = 105.0 Å and a = 95.8, b = 107.6, c = 104.8 Å, respectively, indicating that one asymmetric unit contains two molecules with a VM value of 2.58 (Å3/Da). All data were processed using the HKL2000 package (23), and statistics are shown in Table 1.
Table 1.
Data collection statistics
| Crystal | Se-Met |
Native | ||
|---|---|---|---|---|
| Dataset | Peak | Edge | Remote | |
| Beamline | BL41XU (SPring-8) | BL6A (PF) | ||
| Resolution (Å) | 50–2.07 (2.14–2.07) | 50–2.07 (2.14–2.07) | 50–2.05 (2.12–2.05) | 50–2.05 (2.12–2.05) |
| Space group | C2221 | |||
| Unit cell (Å) | a = 96.0, b = 107.3, c = 105.0 | a = 95.8, b = 107.6, c = 104.8 | ||
| Wavelength (Å) | 0.9793 | 0.9796 | 0.9000 | 1.0000 |
| Unique reflections | 33233 (3123) | 33256 (3161) | 34231 (3377) | 33895 (3383) |
| Completeness (%) | 99.6 (96.8) | 99.5 (96.2) | 99.5 (99.8) | 98.7 (99.7) |
| †Rmerge (%)a | 7.5 (38.0) | 6.9 (38.6) | 6.6 (38.3) | 7.4 (38.3) |
| I/σ | 14.7 (2.5) | 16.9 (3.0) | 18.0 (3.5) | 19.6 (5.0) |
| Multiplicity | 7.1 (6.5) | 7.1 (6.3) | 7.2 (7.0) | 5.4 (5.2) |
Values in parentheses refer to the highest resolution shell †Rmerge = Σh Σj | 〈I〉h − Ihj|/Σh Σj Ihj, where 〈I〉h is the mean intensity of symmetry equivalent reflections.
To identify metal ions present in LldR crystals, X-ray absorption fine-structure spectroscopy (XAFS) measurement for cobalt, nickel and zinc atoms was carried out at BL6A of PF (Tsukuba, Japan). The zinc atom absorption edge spectrum was obtained.
Structure solution and refinement
Using Se-MAD data, six of the eight selenium sites were located and initial phases were calculated using the program SOLVE (24). Phase improvement and automated model building were carried out with the program RESOLVE (25). The initial model was built automatically to ∼81%. Structure refinement was performed on the native data. Using the model built on Se-MAD data as a search model, the structural solution for the native crystal was obtained using the program AMoRe (26). Model building and refinement were carried out semi-automatically using LAFIRE (27) with the refinement program CNS (28). After several iterations of LAFIRE, the model was refined to an R factor of 30.6% at the resolution range 20.0–2.05 Å. The difference Fourier electron density map (Fo-Fc) showed a strong peak that seemed to be due to co-purified zinc ions, and this was confirmed by XAFS (data not shown). After water molecules were located, the model was finally refined to an R/Rfree factor of 20.5/25.0% using the program CNS (28), and the stereochemical quality was analysed using the program PROCHECK (29). The refinement statistics for the structure are listed in Table 2.
Table 2.
Refinement statistics
| Refinement | Native |
|---|---|
| Resolution range (Å) | 20–2.05 |
| Total no. of non-hydrogen atoms | |
| Protein | 3524 |
| Water | 400 |
| Zinc | 2 |
| R/Rfree (%)a | 20.5/25.0 |
| RMSD bond length/angle (Å/°) | 0.006/1.108 |
| B factor (Å2) | |
| Protein | 28.9 |
| Water | 38.2 |
| Zinc | 25.6 |
| Ramachandran plot (%) | |
| Most favoured regions | 95.2 |
| Additionally allowed regions | 4.6 |
| Generously allowed regions | 0.2 |
| Disallowed regions | 0 |
aR = ∑ |Fobs − Fcal|/∑Fobs, where Fobs and Fcal were observed and calculated structure factor amplitudes. Rfree value was calculated for R using a random 10% subset from all reflections.
Construction of the lldR knockout strain
Using C. glutamicum ATCC31831 as a wild-type strain, an lldR knockout strain was constructed by a two-step homologous recombination. A 2150-bp DNA fragment encompassing the lldR gene was amplified by PCR using the primer pairs ΔlldR-fwd/rev (GGCGTCCACTCTAGATTCTGCGAAGC (XbaI)/TGGGCATGCGAGCGTTGAATCGTACGG (SphI) (restriction sites for digestion are underlined, and mutated nucleotides are indicated in bold and italic type; the same convention is used for the primers below). The PCR product was digested with XbaI and SphI and cloned into the plasmid pK18mobsacB (30) digested with the same enzymes. From the resulting plasmid, a PstI fragment (318 bp) containing an internal part of the lldR gene was removed by digestion with PstI followed by self-ligation. The constructed plasmid, named pK18ΔlldR, was transferred into the wild-type strain ATCC31831 and selected by kanamycin resistance. As the plasmid pK18 ΔlldR did not have a replication origin functional in C. glutamicum, only cells in which homologous recombination occurred between the chromosomal DNA and plasmid DNA could grow on plates containing kanamycin. Hence, transformants with kanamycin resistance carried the intact lldR gene and the deleted lldR gene, including the plasmid sequence on their chromosome. One transformant was inoculated into L liquid medium containing no kanamycin, and grown at 30°C overnight. The cells were then spread on L agar plates supplemented with 20% sucrose. As cells carrying the sacB gene encoded by the plasmid DNA that had been integrated into the chromosome were killed in the presence of sucrose, only those cells in which the sacB gene had been excised from the chromosome by a second homologous recombination between the intact and deleted lldR could grow on the plates supplemented with sucrose. From sucrose-tolerant recombinants, the desired lldR knockout strain was selected by PCR amplification with the primers used for cloning of the lldR gene. The selected strain with an lldR knockout was further confirmed by Southern hybridization (data not shown). The lldR knockout strain was named ΔlldR.
Total RNA extraction and primer extension analysis
ATCC 31831 (wild-type) and ΔlldR (lldR knockout) were grown in L liquid medium at 30°C. Two volumes of RNAprotect Bacterial Reagent (Qiagen, Valencia, CA, USA) were added directly to one volume of exponentially growing cultures at an OD660 of ∼0.5 (in logarithmic phase) to stabilize cellular RNAs. The cells were harvested by centrifugation at 5500 g for 15 min at 4°C and total cellular RNAs were isolated using an RNeasy Midi Kit (Qiagen). The isolated RNA was aliquoted and stored at −80°C for later use.
The transcriptional initiation start site of the cgl2917-lldD operon was determined by nonradioactive primer extension analysis. Aliquots of 50 µg of total RNA extracted from the ATCC31831 and ΔlldR strains were used for primer extension with Superscript II RNase H-reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and the biotinylated oligonucleotide cgl2917-ex (CGGTGAGTTTTGATCTACTGCGG), which was complementary to a region within cgl2917. The primer extension products were separated on a denaturing polyacrylamide gel (8-M urea, 6% gel) together with sequencing ladders obtained using the same primer. The template for the sequencing ladders was prepared using the following procedure. A PCR-amplified DNA fragment encompassing cgl2917 was obtained using the primer pair cgl2917-fwd (AATGGGGGGAATTCGATTCGACTGTTTTCC (EcoRI) and cgl2917-rev (AGTAGTTCTGCAGGGTTGGGCAGTTGACGTT (PstI)). The target DNA fragment was cloned into the C. glutamicum/E. coli shuttle vector pECt and transformed into E. coli JM109. Finally, for template use, the plasmid was extracted from 1.5-ml overnight cultures of the cells with a Plasmid Midi Kit (Qiagen). Separated products were detected by chemiluminescence using a Phototope-Star detection kit (New England Biolabs Inc., Beverly, MA, USA).
DNA microarray analyses
To determine changes in the patterns of gene expression due to the absence of the lldR gene, mRNA levels for the two strains ATCC 31831 and ΔlldR were compared by DNA microarray analyses. The C. glutamicum-specific microarray NimbleGen Systems (Madison, WI, USA) (31) includes 2993 open reading frames (ORFs) from the C. glutamicum ATCC 13032 genome. Each ORFs was represented by 32 probe pairs of 24-mer oligonucleotides, each pair consisting of a sequence perfectly matched to the ORF and another adjacent sequence that harbours two mismatched bases for determination of background and cross-hybridization. Synthesis of fluorescently labelled cDNA from total RNA, microarray hybridization and data analysis were carried out by NimbleGen Systems based on standard procedures (31). For statistical data analysis of the gene expression data, the P-value for each ORF was calculated using Student's t-test. ORFs with relative changes of at least 1.5, P < 0.05 and present on two microarrays were considered significant.
DNase I footprint analysis
The plasmids containing ∼110 bp upstream and downstream of the binding sites were first constructed by TA cloning with the pGEM-T Easy Vector (Promega), and the primer sets cgl2917F-fwd/rev (GTCATGCAGATTAACTAACTC/GCTTGTTGAGTTGCTGCTTG) and cgl1934F-fwd/rev (CATACGGTTCTTCCACAATC/GAAGATCACGTCGCACTG) were used for footprint analysis of the genes cgl2917 and cgl1934, respectively. Subsequently, the probes for DNase I footprint experiments were prepared by PCR from the constructed plasmids. Prior to this PCR amplification, the 5′ termini of the above primers of either the coding or the noncoding strand were labelled with [γ-32P] ATP (PerkinElmer Life Sciences, Boston, MA, USA) using T4 polynucleotide kinase (Takara Bio, Otsu, Japan). Aliquots of 120 ng of labelled probes, 10 µg of salmon testis DNA (Sigma) and the purified LldR protein (final protein concentration 15 nM) were mixed in 20 μl of reaction buffer (40 mM Tris–HCl, pH 7.5, 200 mM NaCl, 8 mM MgCl2, 5 mM dithiothreitol) and incubated for 20 min at 37°C. After incubation, 0.25 U of DNase I (Takara Bio) in 0.5 μl of reaction buffer were added and mixed, and digestion was allowed to progress for 1 min at 37°C. The digestion reaction was terminated by addition of phenol/chloroform, and reaction mixtures were extracted. DNA was precipitated with ethanol, and the resulting pellets were re-suspended in 8 μl of loading dye. After heating at 90°C for 1 min, aliquots were analysed by electrophoresis on an 8-M urea-containing 6% polyacrylamide gel. Sequence ladders were generated using a Thermo Sequence Cycle Sequence Kit (Amersham Biosciences) with the same primer end-labelled for probe preparation.
Isothermal titration calorimetry (ITC)
ITC measurements were performed at 25°C using a VP-ITC MicroCalorimeter (MicroCal Inc., Northampton, MA, USA). In the ITC experiments, the protein and DNA samples were prepared with buffer containing 20 mM Na-Pi, pH 8.0, 0.2 M NaCl, 0.5 mM EDTA and all sample solutions were filtered and degassed. Titration was carried out using a 370-µl syringe with stirring at 307 r.p.m.; each titration consisted of an initial 3-µl injection, followed by 24 subsequent injections of 10 μl into the ITC cell (∼1.4 ml). For ITC measurements, synthetic oligodeoxyribonucleotides were used (cgl2917-site1 DNA-F/R, TTGTGGTCTGACCATGA/TCATGGTCAGACCACAA; cgl2917-site2 DNA-F/R, AGGTTGGGCCAATCGGTT/AACCGATTGGCCCAACCT; cgl2917-site1–2 DNA-F/R, TTGTGGTCTGACCATGAGGTTGGGCCAATCGGTT/AACCGATTGGCCCAACCTCATGGTCAGACCACAAGAA; cgl1934-site DNA-F/R, TAAGCATGGTCAGCCAAACGGAA/TTCCGTTTGGCTGACCATGCTTA; Hokkaido Biosystems, Sapporo, Japan). Equimolar amounts of the complementary strands were mixed in annealing buffer (20 mM Na-Pi, pH 8.5, 0.2 M NaCl, 0.5 mM EDTA). Double-stranded DNA (dsDNA) was prepared by annealing the DNA mixture, heating the reaction at 90°C for 5 min, then slowly decreasing the temperature to 4°C overnight. All dsDNA solutions were contained in the ITC cell at a concentration of 10 μM, and the concentrations of the titrated LldR dimer in the syringes for cgl2917 and cgl1934 site DNAs were 200 and 100 μM, respectively. To correct the dilution and mixing effects, blank titrations were conducted by injecting protein solution into the cell containing only buffer under the same conditions, and subtracted from the raw data. All data were analysed with MicroCal Origin 7.0 (Microcal Software, Inc., MA, USA).
Electrophoretic mobility shift assay (EMSA)
The dsDNA (cgl2917-site1) was prepared in the same way as in ITC experiments. The LldR protein with a single-residue mutation was mixed with the dsDNA in binding buffer (same as buffer B in protein preparation), and the protein–DNA mixture was incubated for 1 h at room temperature. The incubated samples were separated on native polyacrylamide gels in TBE buffer at 4°C and a constant current of 8 mA. The gel was finally stained with ethidium bromide, photographed with LAS-3000 mini (Fujifilm, Tokyo, Japan) and analysed with Multi Gauge Ver 3.0 software (Fujifilm, Japan).
RESULTS
Overall structure
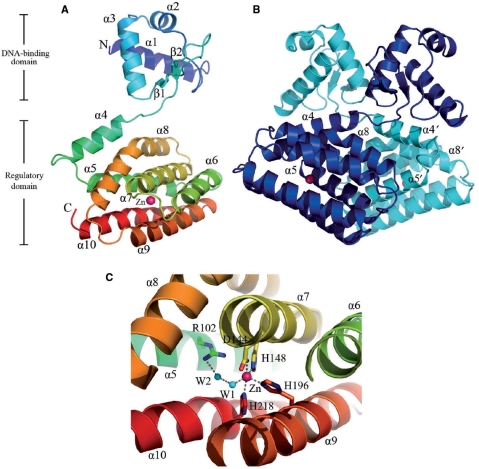
The crystal structure of LldR was determined using the Se-MAD method, and the structure (Figure 1) was refined to 2.05-Å resolution using native data. The final model contains two monomers (monomer A, Ser2–Leu232; monomer B, Ser2–Ala231) (Figure 1B), two zinc ions and 400 water molecules in an asymmetric unit, and has been deposited in the Protein Data Bank (code 2DI3). The two copies were well superimposed with a root mean square deviation (r.m.s.d.) of 1.3 Å on Cα atoms except for residues 58–72, which form an anti-parallel β-sheet and have distinct conformations (the maximum and average distances between Cα atoms are 13.1 and 6.3 Å, respectively). The two monomers form a dimer, which is consistent with the results of gel filtration experiments (data not shown).
Figure 1.
The crystal structure of LldR in complex with co-purified Zn2+. (A) Ribbon representation of the LldR-Zn2+ complex in a monomer. The ribbon model is coloured according to the sequence from blue at the N-terminus to red at the C-terminus, and the co-purified zinc ion is shown as a pink ball. (B) Ribbon representation of the LldR-Zn2+ dimer. The two monomers are coloured cyan and blue. (C) Close-up view of the Zn2+ binding site. The amino acid residues and a water molecule binding to Zn2+ are shown as sticks and a cyan ball, respectively. The figures were produced using PyMOL (DeLano Scientific LLC, http://pymol.sourceforge.net).
The monomer structure is composed of separated N- and C-domains (Figure 1A), including 10 α-helices and two β-strands. The N-domain with the topological order α1, α2, α3, β1 and β2, contains a typical prokaryotic winged HTH DNA binding motif (32). This motif is formed by helices α2, α3 and their connecting turn, together with a winged anti-parallel β-sheet (Figures 1A and B, and 2). The C-terminal region (Gly77–Leu232), consisting of seven helices (α4–α10) forming a helical bundle, is assumed to act as a regulatory domain with a role in dimerization and ligand binding. There is a severe kink at Arg168 in helix α8, resulting in the formation of a helix with a vaulted bridge shape that crosses the bundle. The bundle core contains mainly aromatic and aliphatic side chains, except for several residues with charged side chains (His, Asp), which will be described later.
Figure 2.
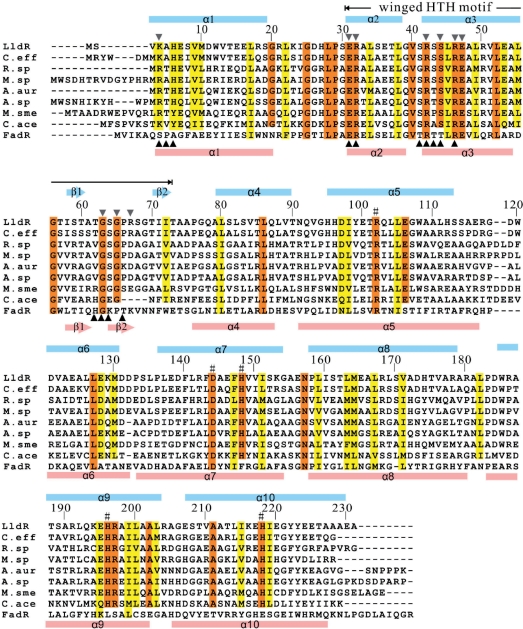
Sequence alignment of LldR homologues and FadR. Sequences are numbered according to that of LldR. The conserved and conserved-changed residues are indicated by red and yellow shaded boxes, respectively. The residues involved in Zn2+ binding in LldR are indicated by #. Black triangles, downward and upward, indicate the residues contacting DNA in LldR and FadR, respectively. The secondary structures (arrows represent β-strands; bars represent α-helices) of LldR and FadR are coloured cyan and pink, respectively. The LldR homologues are: C. eff (Corynebacterium efficiens), NP_739367; R. sp (Rhodococcus sp. RHA1), YP_703439; M. sp (Mycobacterium sp. JLS), ZP_01275280; A. aur (Arthrobacter aurescens), YP_949019; A. sp (Arthrobacter sp. FB24), YP_832817; M. sme (Mycobacterium smegmatis), YP_885298; C. ace (Clostridium acetobutylicum), NP_349157.
Approximately 3993 Å2 of accessible surface area was buried upon dimerization, with 17% of the total surface area of each monomer. The contacts are mainly formed by side chains (mainly Leu) along helices α4, α5 and α8, and their linked loops including Ala79–Leu55′, Leu80–Thr162′/Leu163′, Leu86–Leu17′/Ala74′, Val89–Thr14′/Arg17′, Gln92–Ile24′, Leu104–Leu104′/Trp108′, Asn157–Asp97′/Glu100′, Leu159–Val93′/Ile98′, Leu163–Leu80′/Thr101′/Leu167′ and Leu167–Leu80′/Leu159′, where the prime refers to the second monomer in the dimer. In addition, the interactions Glu47–Arg50′ and Val51–Ala54′ through helix α3 in the N-domain also contribute to dimer formation. The interactions in the N-domain are much weaker than those in the C-domain. Of the residues involved in dimer formation, Leu17, Gln47, Leu55, Leu80, Leu86, Asp97, Ile98, Leu159, Leu163 and Leu167 are conserved or conserved-changed among LldR homologues and FadR (Figure 2).
Although Zn2+-containing buffer was not used during purification and crystallization, a co-purified zinc ion was assigned to each LldR molecule in the final structure based on both the electron density maps and XAFS check. The zinc ion was refined well at the binding site with an average B factor of 25.6 Å2, which was comparable to those of the surrounding atoms. The zinc ion was located at the core of the helical bundle with a coordination number of five (NE2 of His148, His196 and His218; OD1 of Asp144; water molecule W1) (Figure 1C). The distances between the zinc ion and the five coordinated atoms in the two monomers ranged from 2.1 to 3.1 Å. This zinc ion interaction network was involved in four helices of the helical bundle, which presumably assists in stabilizing the structure. The present structure first revealed that a protein belonging to the GntR family has a strong ability to bind a zinc ion, although the detailed biofunction of the zinc ion is not yet clear. The residues involved in Zn2+ binding are completely conserved in LldR and its homologues (excluding FadR), indicated by the mark # in Figure 2, revealing a common structural feature of Zn2+ binding in the regulatory domain. LldR and its homologues may compose another subfamily of the GntR family, differing from the FadR subfamily in this structural feature.
Structural comparison
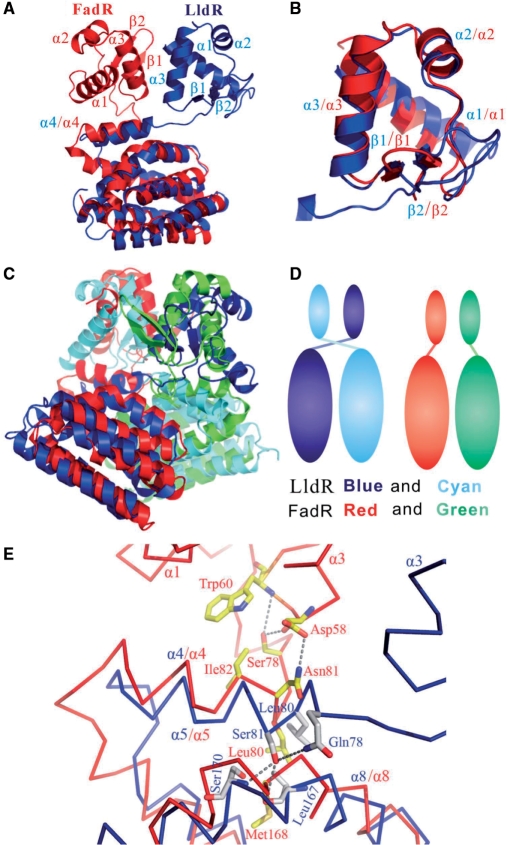
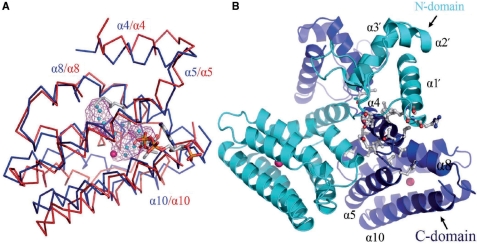
A Dali Server (33) search showed that the LldR protein has the greatest structural similarity to FadR (34) in the apo form (PDB: 1E2X) with a Z-score of 16.5. FadR is a fatty acid-responsive transcription factor in E. coli, which plays significant roles in balancing the anabolic and catabolic fatty acid pathways (35,36). Both LldR and FadR contain two domains (N- and C-domains), and both N- and C-domains of the two proteins were superimposed well with respective r.m.s.d values of 1.4 and 1.8 Å on Cα atoms by the program LSQKAB (37) (Figure 3A and B). The dimeric structure of LldR also resembles that of FadR, as shown in Figure 3C. However, when the C-domain of LldR was superimposed with that of FadR, the N-domains of the two proteins maintained distinct positions in relation to the C-domains (Figure 3A), indicating that the domains are assembled in different orientations. Thus, overall similarity of the two dimeric structures was achieved by domain swapping (Figure 3D); when the C-domains of monomers A and B in the LldR dimer were superimposed with those in the FadR dimer, the corresponding N-domains of monomers A and B were exchanged between the two structures.
Figure 3.
Structural comparison of LldR and FadR (apo form). (A) Structures of LldR and FadR monomers after superposing C-domains. LldR and FadR are shown as ribbon representations coloured blue and red, respectively. (B) Structural superposition of DNA-binding domains of LldR (blue) and FadR (red). (C) Structural superposition of LldR and FadR homodimers. The two monomers are coloured blue and cyan in LldR and red and green in FadR. (D) Schematic diagram showing domain swapping for LldR and FadR in homodimer assembly. (E) Enlarged view showing a comparison of interactions between the α4-linker and the domains for CGL295 and FadR. Cα traces of LldR and FadR are coloured blue and red, respectively; the residues are shown in a stick model, with oxygen atoms in red, nitrogen atoms in blue and carbon atoms grey in LldR and yellow in FadR.
Domain swaps may be due to swapping of small fragments, swapping of structural and functional motifs and recombination of domains in multidomain proteins (38). The mechanism of domain swapping is believed to be important for understanding the evolution of proteins (38,39). To our knowledge, this is the first observation of domain swapping in a dimeric transcription factor. An enlarged view of the interactions between the linker and the connected domains of the two structures is shown in Figure 3E. When the C-domain of LldR is superimposed with that of FadR, the two N-domains diverge at residue 81 (Ser81 in LldR and Asn81 in FadR) located at the N-terminal side of helix α4, leading to distinct orientations of the respective N-domains (Figure 3A and E). The residue interactions are quite different in the diverging regions. Hydrophobic interaction between Leu82 with Leu55′ is important to change the trajectory of the domain linker. The residues in the domain linker of FadR make extensive contacts with those of the N-domain, including Asn81–Asp58 (3.1 Å), Ser78–Asp58 (2.92 Å), Ser78–Trp60 (3.96 Å) and Ile82–Trp60 (∼4 Å) (Figure 3E). In contrast, no interaction was observed between the domain linker and the N-domain within the LldR monomer. Obviously, the interactions between the domain liker and domains are associated with the different arrangements of the N- and C-domains for the two protein monomers.
Comparison of the expression profiles of C. glutamicum wild-type and ΔlldR mutant using DNA microarrays
The helix–turn–helix (HTH) GntR family of transcription factors comprised of FadR, HutC, MocR, YtrA, AraR and PlmA subfamilies, regulates varied biological processes (34,40–44). The FadR subfamily grouping 40% of GntR-like regulators is the largest, and most of its members are involved in the regulation of enzymes with roles in substrate oxidization related to amino acid metabolism (40). This function is important for C. glutamicum, which is the most efficient producer of amino acids in industrial applications. Structural analysis and sequence alignment showed that LldR and FadR have a similar dimer structure (functional unit) and DNA-binding motif residues (Figures 1 and 2). FadR regulates at least 15 genes and operons (35,36), suggesting that LldR may also be an important transcription factor. To investigate the effects of LldR on global gene expression and to find the predicted target genes, genome-wide DNA microarray analyses were performed by comparing mRNA levels of the two strains ATCC31831 and ΔlldR. The results are shown in Table 3.
Table 3.
Relative mRNA levels based on a comparison of global gene expression in wild-type (WT) and lldR knockout (ΔlldR) strains
| Gene (cgl no.) | Function | Ratio (ΔlldR/WT)a |
|---|---|---|
| 0370 | Succinate dehydrogenase (Sdh) subunit C | 0.55 |
| 0371 | Sdh subunit A | 0.58 |
| 0372 | Sdh subunit B | 0.66 |
| 1578 | 6-Phosphogluconolactonase | 0.65 |
| 1931 | Transcriptional regulator | 0.63 |
| 1934 | Transcriptional regulator of sugar metabolism | 2.14 |
| 1935 | Fructose-1-phosphate kinase (FruK) | 1.97 |
| 1936 | PTS system, fructose-specific EII component | 2.06 |
| 2381 | Transcriptional regulator | 0.58 |
| 2541 | Phosphoglucomutase | 0.59 |
| 2642 | PTS system, sucrose-specific EII component | 2.24 |
| 2911 | l-lactate dehydrogenase (cytoplasm) | 1.63 |
| 2917 | Putative integral membrane transport protein (permease) | 2.12 |
| 2918 | l-lactate dehydrogenase | 2.18 |
Genes of which the mRNA levels were changed by >1.5-fold (P < 0.05, Student's t-test) are listed.
aThe mRNA ratios represent average values obtained from two DNA microarray experiments in which RNA was isolated from three independent cultures in L broth medium at 30°C.
In this investigation, 14 genes listed in Table 3 fulfilled the experimental criterion, i.e. that the mRNA levels of the two strains differ by more than 1.5-fold (P < 0.05; Student's t-test). Of these 14 genes, half showed a higher mRNA level and half a lower level after lldR knockout. With the exception of three genes encoding transcriptional regulators, of which only one has a known role in sugar metabolism, the rest have functions related to carbohydrate metabolism. In particular, the levels of expression of all the genes in cgl2917-cgl2918 and cgl1934-cgl1935-cgl1936 operons in ΔlldR were approximately double those in wild-type controls. In contrast, the mRNA levels of three genes, cgl0370, cgl0371 and cgl0372, which form an operon encoding succinate dehydrogenase C, A and B subunits, respectively, were decreased by 0.55-, 0.58- and 0.66-fold, respectively. It is likely that transcription factor LldR also regulates these genes either directly or indirectly.
Identification and characterization of LldR-binding site on DNA
To confirm the direct target genes of the transcription factor LldR, its binding site on DNA was identified and characterized. The FadR dimer binds a pseudopalindromic 17-bp DNA sequence that shows the highest affinity for the binding site in the fadB gene, which has more precise dyad symmetry containing the consensus motif TGGTN3ACCA (34–36). The residues Arg35, Arg45, Arg49 and Gly66 in FadR, which are indispensable for DNA binding, are completely conserved in LldR (Figure 2). These observations suggest that LldR binds to DNA in a manner similar to FadR. Therefore, for the above genes, two qualifications were used for screening: first, the DNA covering 500 bp upstream and 100 bp downstream of the translational start site was chosen for analysis using the online program NNPP (Promoter Prediction by Neural Network for Prokaryotes or Eukaryotes, http://www.fruitfly.org/seq_tools/promoter.html); second, the binding site should be ∼17 bp in length with partial dyad symmetry (palindromic or pseudopalindromic sequence) overlapping or downstream of the predicted promoter, and contain a consensus sequence similar to 5′-TGGTNxACCA-3′.
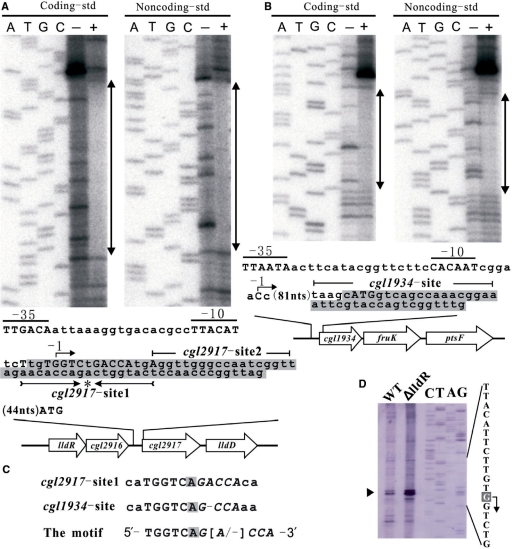
As a result, the candidate binding sites with regard to the cgl2917-lldD and cgl1934-fruK-ptsF operons were obtained. The binding affinities of LldR to these sites were preliminarily verified by EMSA on dsDNA produced from the synthetic complementary oligonucleotides (data not shown). Subsequently, the results of DNase I footprint analyses indicated that LldR protein at a concentration of 15 nM protects DNA covering from nucleotide −4 to +29 (coding strand) and –7 to +25 (noncoding strand) of cgl2917, +87 to +105 (coding strand) and +83 to +101 (noncoding strand) of cgl1934, respectively. Therefore, the 32–33-bp sequences in cgl2917, and 19-bp sequences in cgl1934 were confirmed to be the two binding sites for LldR. The location of the predicted promoter for the cgl2917-lldD operon was consistent with the results of primer extension analysis of total RNAs isolated from the strains ATCC31831 and ΔlldR grown on LB medium, which demonstrated that the transcriptional start site (defined as –1) is located 74 bp upstream of the translational initiation ATG codon of the cgl2917 gene with two sequences TTACAT and TTGACA in the promoter defined as −10 and −35 regions, respectively (Figure 4A and D). Primer extension analysis further demonstrated that the amount of cgl2917 transcript (indicated by black triangle in Figure 4B) in the ΔlldR cells was 2.7-fold higher than that in wild-type controls, in agreement with the results of DNA microarray analyses (Table 3). These results taken together indicated that LldR functions as a transcriptional repressor to directly control the cgl2917-lldD operon. In the case of the cgl1934 (Figure 4B), the putative transcriptional start site (C, defined as –1) is situated 88 bp upstream of the translational initiation ATG codon; accordingly, the two sequences CACAAT and TTAATA were designated as −10 and −35 regions. The LldR-binding region is more than 82 bp downstream of the transcriptional start site, implying that LldR is also a complicated repressor regulating the cgl1934-fruK-ptsF operon.
Figure 4.
Footprint analysis of LldR protein. (A) Analysis in the promoter regions of genes cgl2917. (B) Analysis in the promoter regions of cgl1934. The protected areas of LldR protein at a concentration of 15 nM obtained from the footprint experiments are indicated by the shaded box in the schematic representation of the promoter regions of genes cgl2917 and cgl1934. With the exception of the protected areas, only the coding strand is shown, and the translational initiation codons of the genes cgl2917 and cgl1934 are indicated in upper case (ATG). The putative transcriptional start indicated by the arrow was defined as –1. The cgl2917-site1 contains an evidently inverted repeat sequence indicated in upper case, with the centre of symmetry marked by an asterisk (*). (C) Representation of the consensus sequence. The two sequences with high similarity and the functional motif are shown. The centre of symmetry and the half-site of the inverted repeat are marked by the shades box and shown in italic type, respectively. (D) Primer extension analysis of the cgl2917 gene. The transcriptional initiation base is shown in white on a grey background and indicated with an arrow. The ratio between wild-type and ΔlldR strains of the levels of cgl2917 mRNA, indicated by the black triangle, was 2.7.
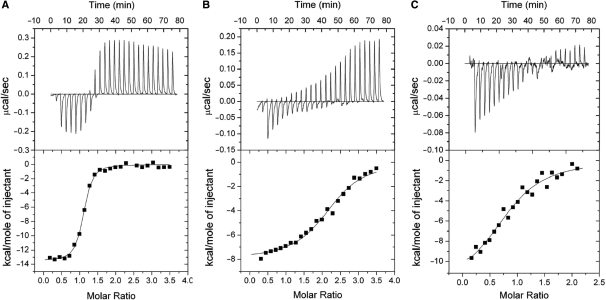
The lengths of the two LldR-binding sequences are quite different as determined by footprint analyses. Taking the size of the LldR protein into account, it seems that two LldR dimers bind to the long sequence of cgl2917 (cgl2917-site1–2, 34 bp), and one dimer to the sequence of cgl1934. Four types of dsDNA representing the full-length (cgl2917-site1–2) or the two halves (cgl2917-site1, cgl2917-site2) of cgl2917, or the cgl1934-site (Figure 4A and B), were thus prepared to perform ITC experiments. The results of ITC showed that the stoichiometry of protein and DNA was clearly one LldR dimer to cgl2917-site1 and cgl1934-site, and two LldR dimers to cgl2917-site1–2, corresponding to Kd values of 8.1 × 10 nM, 1.1 × 103 nM and 1.7 × 103 nM, respectively (Figure 5). The Kd values indicated that LldR has the highest affinity for cgl2917-site1, which contains an inverted repeat sequence with more precise symmetry (5′-−4TtgTGGTCTGACCAtgA+13-3′, the centre of symmetry and the half-site of the inverted repeat are underlined and shown in italic type, respectively) (Figure 4A and D). The typical fitting curve was not observed for the titration of LldR to cgl2917-site2-dsDNA, implying a weak LldR–DNA interaction, which was consistent with the results of in vitro binding assay by EMSA (data not shown). These results, taken together, suggest that binding of one LldR dimer to cgl2917-site1 enhances binding of the second dimer to cgl2917-site2, but the close proximity of the two dimers binding decreases the affinity for cgl2917-site1, with Kd from 8.1 × 10 nM to 1.1 × 103 nM.
Figure 5.
ITC of the interaction of LldR and DNA. Representative plots from ITC experiments are illustrated with raw data in the upper panel and fitting curves (continuous lines) in the lower panel for (A–C). The concentrations of titrated LldR dimer were 200 (A and B) and 100 μM (C). The thermodynamic values calculated by curve fitting were: A (cgl2917-site1 dsDNA), ΔS = –14.1 cal/mol, ΔH = –13.9 ± 0.1 kcal/mol, KB = 1.2 ± 0.2 × 107 M–1 (Kd = 8.1 × 10 nM), N = 1.1 ± 0.01; B (cgl2917-site1–2 dsDNA), ΔS = 0.58, ΔH = –8.0 ± 0.2 kcal/mol, KB = 9.1 ± 1.4 × 105 M–1 (Kd = 1.1 × 103 nM), N = 2.2 ± 0.03; C (cgl1934-site dsDNA), ΔS = –15.1 cal/mol, ΔH = –12.4 ± 1.2 kcal/mol, KB = 5.7 ± 1.6 × 105 M–1 (Kd = 1.7 × 103 nM). N = 0.9 ± 0.05. N is the stoichiometry of bound LldR dimer per dsDNA, and the dissociation constant Kd is the reciprocal of the affinity constant KB (Kd = 1/KB). Note that the initial injection was 3/10 volume of the subsequent injection.
Ala-scanning mutation and DNA binding
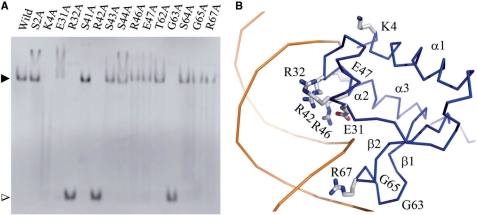
To determine the residues that interact with DNA, the mutated LldR proteins and the cgl2917-site1 DNA were used to examine DNA-binding ability (Figure 6A). Combining sequence and structural comparisons of LldR with DNA-bound FadR (45,46) (Figure 2), 15 residues in LldR were selected and mutated to Ala. The results of EMSA indicated that four residues, Lys4, Arg32, Arg42 and Gly63, are crucial for binding to cgl2917-site1, and substitution of any of these residues with Ala results in complete loss of DNA binding. Decreases in DNA binding were observed in the cases of E31A, R46A, E47A, G65A and R67A. Taken together, these observations indicated that the residues Lys4, Glu31, Arg32, Arg42, Arg46, Glu47, Gly63, Gly65 and Arg67 in LldR are important for DNA binding. With the exception of Arg67, all of these residues are conserved or show conservative changes in LldR and its homologues (Figure 2). Moreover, most of these residues are distributed in the same DNA binding regions as those of FadR, indicated by black triangles in Figure 2.
Figure 6.
DNA-binding assay and DNA-binding model of LldR. (A) Results of in vitro DAN-binding assay of LldR and its mutants. All proteins (native and mutated) were purified according to the same protocol under similar conditions. Site 1 DNA was used; the amounts of dimeric protein and dsDNA were 20 and 8 pmol, respectively. The LldR–DNA complex and free DNA bands are indicated by black and white triangles, respectively. The images were analysed with Multi Gauge Ver 3.0 software (Fujifilm). (B) Model of DNA and protein contacts. Only the DNA-binding domain of one monomer is presented, shown as a Cα trace. The residues contributing to contact with DNA, as determined by ALA-scanning mutation, are shown as stick models for side chains, with carbon, oxygen and nitrogen atoms in grey, red and blue, respectively. The DNA is shown as a double-stranded model coloured orange.
Combining the results of ALA-scanning mutation and structural comparison, a model of LldR–DNA was constructed by overlapping LldR onto the FadR–DNA complex (Figure 6B). The paired HTH motifs (α2–turn–α3) contact DNA by projecting into the major groove, and the two winged β-hairpins dock into the flanking minor grooves. The residues that interact with DNA could be grouped into four regions (marked by black downward triangles in Figure 2). At the N-terminus, residue Lys4 makes a significant contribution to DNA binding. Similar to the case in FadR, this binding may be non-specific, through the interaction between the side chain of Lys4 and the phosphate backbone (45,46). The second interaction region is located at the beginning of helix α2, and is composed of Glu31 and Arg32. Such a binding mode is very common in the HTH family of transcription factors. The residue Arg32 may project into the major groove of the DNA helix and make a specific interaction with the DNA base. The turn between α2 and α3 and the tip of helix α3, including Arg42, Ser44, Arg46 and Glu47, forms the third contact region. With the exception of Ser44, mutating any of these residues to Ala (particularly Arg42) resulted in a decrease of complete loss of DNA binding. It is likely that Arg42 as well as Arg32 in LldR donate specific hydrogen bonds to acceptors of DNA bases in the major groove, in a manner similar to that of residues Arg45 and Arg35 in FadR (45,46). In HTH proteins, helix α3 is termed ‘the recognition helix’, as it is inserted into the major groove of the DNA helix and is critical for specificity (47). The final interaction region is composed of the residues Gly63, Gly65 and Arg67, which lie in the tip of the winged β-hairpin interacting with the minor groove of the DNA helix. Of these residues, the conserved residues Gly63 and Gly65 are crucial for LldR–DNA contact. Similar to FadR, these Gly residues may save space for the tip of the wing to approach the minor groove of the DNA helix.
DISCUSSION
The putative ligand binding-cavity and switch mechanism
The HTH GntR family has about 270 members distributed among the most diverse bacterial groups, which regulate a wide variety of biological processes. In general, proteins belonging to this family contain an N-terminal DNA-binding domain and a C-terminal ligand-binding/dimerization domain (also termed a regulatory domain). The C-domain of the FadR subfamily is composed of an α-helical bundle in most cases (40). The C-domains of FadR in the apo, DNA binding and ligand acyl-CoA binding (excluding helices α4 and α8) forms show similar conformations (45). By considering the similarity of the C-domain conformations of FadR in three forms, and between LldR and FadR, C-domain comparison could provide insight into the ligand-binding site of LldR. As shown in Figure 7, myristoyl-CoA penetrates the seven-helix bundle of FadR through entry between helices α5 and α10, and the C14 acyl chain is buried deep inside the C-domain cavity, while several water molecules are buried deep inside this cavity in the apo form (34,45). LldR has a similar C-domain cavity filled with 11 water molecules and one zinc ion (Figure 7A). This cavity has an estimated volume of 158 Å3 (calculated using a program developed by our group), which is comparable with that of FadR in apo form (142 Å3) (34). The cavity of LldR is surrounded by residues Ile98, Arg102, Leu105 and Glu106 in α5, Leu141, Asp144, Val145, His148 and Val149 in α7, Met164, Leu167, Val171, His174 and Thr175 in α8, Leu192 and His196 in α9, and His218 and Tyr222 in α10. With the exception of Arg102, Asp144, His148, His196 and His218, which are involved in the zinc ion interaction network, most of these residues have aliphatic or aromatic side chains (hydrophobic residues), suggesting that the ligand would bind to LldR predominantly through hydrophobic interaction with these residues. Combining structural features of the cavity and the nature of the regulatory genes, it can be postulated that the ligand would contain a carbon chain, as found in lactate, fructose, etc. Although, binding of these compounds to LldR could not be detected by co-crystallization or crystal soaking, it was recently reported that l-lactate interfered with binding of LldR to its target DNA (48).
Figure 7.
Structural comparison of LldR and FadR–acyl–CoA complexes. (A) Comparison of the ligand-binding domains of LldR and FadR–acyl–Co complex. The structures of LldR and FadR–acyl–CoA complexes are shown as a Cα trace, coloured blue and red, respectively. The acyl–CoA is represented as a stick model. In LldR, the cavity is shown as a magenta chicken wire structure, and water molecules and the zinc ion occupying the cavity are shown as cyan and pink balls, respectively. (B) Putative switch region of LldR. The two monomers are coloured blue and cyan. The contacting residues between helix α4 and the boundary of the putative ligand-binding cavity, and between helix α4 and the DNA-binding domain (N′-domain), are shown as a stick model.
The entrance to the cavity in LldR seems the same as that of FadR (Figure 7A). In FadR, ligand binding to the cavity in the C-domain triggers conformational changes of two residues, Met168 and Tyr172 in helix α8 surrounding the cavity, which causes a shift of helix α4 toward helix α1 in the N-domain. This shift results in movement of the DNA-binding domain and consequently decreases the binding affinity for DNA (45). In the case of LldR, ligand binding to the aforementioned cavity (Figure 7A and B) can be hypothesized to also trigger conformational changes, which would introduce a shift of helix α4, resulting in motion of the N′-domain (the prime refers to another monomer in the homodimer) through contacts between helix α4 and the N′-domain. The positions of the paired N-domains would thus be changed, resulting in a decrease of DNA-binding affinity. In both proteins, helix α4 appears to be a key component in conformation transmission. While helix α4 of FradR makes extensive interactions with the N-domain within a monomer (45,46), helix α4 in LldR contacts the C-domain and the N′-domain (Thr14′, Leu17′, Arg18′, Ile24′, Leu55′ and Ala74′) by homodimeric assembly. It is obvious that the transmission of ligand-induced conformational change is distinct for the two proteins, in agreement with the apparent swapping of domains during homodimeric assembly.
Function of LldR: a novel transcriptional repressor in control of two operons involved in l-lactate and sugar utilization/re-utilization
The results of the present study indicated that LldR could bind to two DNA regions, overlapping –4 to +29 (coding strand) for the promoter of the cgl2917-lldD operon and +87 to +105 (coding strand) for the putative promoter of the cgl1934-fruK-ptsF operon (Figure 4A and B). Such binding blocks transcription of the two operons, as confirmed by DNA microarray analysis (Table 3). Particularly for the cgl2917-lldD operon, primer extension analysis showed a 2.7-fold increase in mRNA level of the cgl2917 gene (Figure 4C), and SDS–PAGE in combination with mass spectrometry identification also demonstrated a 2.2-fold increase in lldD expression in vivo (data not shown) in the ΔlldR strain. These results, taken together, suggest that LldR functions as a transcriptional repressor for the two operons.
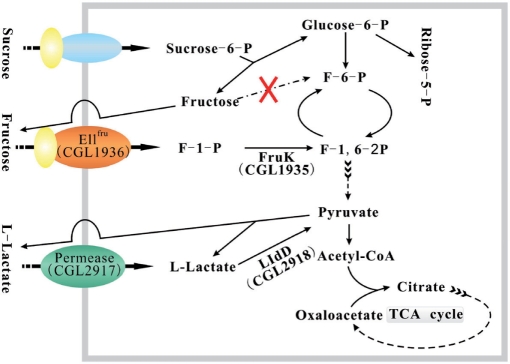
The cgl2917-lldD operon induced during temperature-triggered glutamate production is essential for the utilization or re-utilization of l-lactate (10). The gene cgl2917 encodes a permease that is putatively involved in uptake of l-lactate (Figure 8), and lldD encodes LldD (10). LldD catalyses the oxidation of l-lactate to pyruvate, which can be converted into acetyl-CoA used in the tricarboxylic acid (TCA) cycle (Figure 8). In E. coli, the lld operon involved in l-lactate utilization is controlled by the two-component signal transduction system ArcB/ArcA, and its transcription is increased by l-lactate (49,50). The results of the present study indicated that the lldR gene encodes a repressor regulating the lld operon. Fructose is an important carbon source for industrial fermentation with C. glutamicum. However, regulation of fructose uptake is poorly understood. The present study revealed that the cgl1934-fruK-ptsF operon is also under the control of LldR. This operon encodes a transcriptional regulator of sugar metabolism, FruK and EIIFru (9). EIIFru is a fructose-specific permease involved in converting fructose into F-1-P across the membrane (Figure 8). Subsequently, F-1-P can be converted into F-1,6-2P to enter glycolysis, catalysed by FruK, which is indispensable for growth of C. glutamicum on fructose minimal medium (17). Due to the absence of a fructokinase gene in C. glutamicum (21), fructose from sucrose-6-P (product of membrane transport of the sucrose) can not be converted into F-6-P (Figure 8). Fructose was shown to be excreted, then re-assimilated via a fructose-specific PTS (21,51). Therefore, FruK and EIIFru may form an assisted pathway to utilize intracellular fructose. Thus, LldR plays a significant role in reutilization of both fructose and sucrose by regulating the transcription of the cgl1934-fruK-ptsF operon.
Figure 8.
Schematic diagram representing the functions of the two operons regulated by the transcriptional repressor LldR. The membrane is shown as a rectangular frame coloured grey.
The level of l-lactate excreted as a metabolic by-product was markedly increased when fructose was supplied as the carbon source; however, the reason for this change of carbon flux is not fully understood (6). The results of the present study showed that the affinity of LldR for the operator of the cgl2917-lldD operon is much higher than that of the cgl1934-fruK-ptsF operon. It seems that repression of the latter operon by LldR is weaker and its depression occurs at a lower concentration of inducer as compared to the former, resulting in the re-utilization of fructose prior to l-lactate, the carbon flow from fructose to lactate is highly advantageous for producing lactate in industrial applications. It is necessary to identify the inducer of LldR, which would be helpful to elucidate such a distinction in carbon flux during l-lactate and fructose utilization. The present study is the first step toward an understanding of the regulatory mechanism for l-lactate, fructose and sucrose metabolism, which has a significant role in the control of carbon flux within the central metabolic pathways of C. glutamicum, providing potential strategies for improving amino acid production.
FUNDING
The National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: Grant-in-aid for Scientific Research (No. 19370037) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank M. Takano (Hokkaido University) for technical assistance, and the staff of beamline BL41XU, SPring-8 and beamline BL6A, Photon Factory, Japan, for their help with data collection.
REFERENCES
- 1.Kinoshita S, Udaka S, Shimono M. Studies on the amino acid fermentation. I. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 1957;3:193–205. [PubMed] [Google Scholar]
- 2.Abe S, Takayama K-I, Kinoshita S. Taxonomical studies on glutamic acid-producing bacteria. J. Gen. Appl. Microbiol. 1967;13:279–301. [Google Scholar]
- 3.Hermann T. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 2003;104:155–172. doi: 10.1016/s0168-1656(03)00149-4. [DOI] [PubMed] [Google Scholar]
- 4.Jetten MJ, Sinskey AJ. Recent advances in the physiology, and genetics of amino acid-producing bacteria. Crit. Rev. Biotechnol. 1995;15:73–103. doi: 10.3109/07388559509150532. [DOI] [PubMed] [Google Scholar]
- 5.Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern JL, Cocaign-Bousquet M, Lindley ND. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur. J. Biochem. 1998;254:96–102. doi: 10.1046/j.1432-1327.1998.2540096.x. [DOI] [PubMed] [Google Scholar]
- 6.Kiefer P, Heinzle E, Zelder O, Wittmann C. Comparative metabolic flux analysis of lysine-producing Corynebacteium glutamicum cultured on glucose or fructose. Appl. Environ. Microbiol. 2004;70:229–239. doi: 10.1128/AEM.70.1.229-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda M, Nakagawa S. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 2003;62:99–109. doi: 10.1007/s00253-003-1328-1. [DOI] [PubMed] [Google Scholar]
- 9.Engels V, Wendisch VF. The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 2007;189:2955–2966. doi: 10.1128/JB.01596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, Wendisch VF. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 2005;71:5920–5928. doi: 10.1128/AEM.71.10.5920-5928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendisch VF, de Graaf AA, Sahm H, Eikmanns BJ. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 2000;182:3088–3096. doi: 10.1128/jb.182.11.3088-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocaign-Bousquet M, Lindley ND. Pyruvate overflow and carbon flux within the central metabolic pathways of Corynebacterium glutamicum during growth on lactate. Enzyme Microb. Technol. 1995;17:260–267. [Google Scholar]
- 13.Netzer R, Peters-Wendisch P, Eggeling L, Sahm H. Cometabolism of a nongrowth substrate: L-serine utilization by Corynebacterium glutamicum. Appl. Environ. Microbiol. 2004;70:7148–7155. doi: 10.1128/AEM.70.12.7148-7155.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez H, Cocaign-Bousquet M, Lindley ND. Simultaneous consumption of glucose and fructose from sugar mixtures during botch growth of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 1997;47:600–603. [Google Scholar]
- 15.Mori M, Shiio I. Phosphoenolypyruvate-sugar phosphotransferase systems and sugar metabolism in Brevibacterium flavum. Agric. Biol. Chem. 1987;51:2671–2678. [Google Scholar]
- 16.Kiefer P, Heinzle E, Wittmann C. Influence of glucose, fructose and sucrose as carbon sources on kinetics and stoichiometry of lysine production by Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 2002;28:338–343. doi: 10.1038/sj/jim/7000252. [DOI] [PubMed] [Google Scholar]
- 17.Moon MW, Kim HJ, Oh TK, Shin CS, Lee JS, Kim SJ, Lee JK. Analyses of enzyme II gene mutants for sugar transport and heterologous expression of fructokinase gene in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol. Lett. 2005;244:259–266. doi: 10.1016/j.femsle.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 18.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Mol. Bio. Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaigalat L, Schlueter JP, Hartmann M, Mormann S, Tauch A, Puehler A, Kalinowski J. The DeoR-type transcriptional regulator SugR acts as a repressor for genes encoding the phosphoenolpyruvate:sugar phosphotransferase system (PTS) in Corynebacterium glutamicum. BMC Mol. Biol. 2007;15:104. doi: 10.1186/1471-2199-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka Y, Okai N, Teramoto H, Inui M, Yukawa H. Regulation of the expression of phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS) genes in Corynebacterium glutamicum R. Microbiology. 2008;154:264–274. doi: 10.1099/mic.0.2007/008862-0. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez H, Lindley ND. Complete sucrose metabolism requires fructose phosphotransferase activity in Corynebacterium glutamicum to ensure phosphorylation of liberated fructose. Appl. Envrion. Microbiol. 1996;62:3878–3880. doi: 10.1128/aem.62.10.3878-3880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uy D, Delaunay S, Germain P, Engasser JM, Goergen JL. Instability of glutamate production by Corynebacterium glutamicum 2262 in continuous culture using the temperature-triggered process. J. Biotechnol. 2003;104:173–184. doi: 10.1016/s0168-1656(03)00151-2. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger TC. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr. D. 2003;59:45–49. doi: 10.1107/S0907444902018048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. Sect. A. 1994;50:157–163. [Google Scholar]
- 27.Yao M, Zhou Y, Tanaka I. LAFIRE: software for automating the refinement process of protein-structure analysis. Acta Crystallogr. D. 2006;62:189–196. doi: 10.1107/S0907444905038965. [DOI] [PubMed] [Google Scholar]
- 28.Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Panns NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 30.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 31.Nuwaysir EF, Huang W, Albert TJ, Singh J, Nuwaysir K, Pitas A, Richmond T, Gorski T, Berg JP, Ballin J, et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huffman JL, Brennan RG. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 33.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Aalten DM, DiRusso CC, Knudsen J, Wierenga RK. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J. 2000;19:5167–5177. doi: 10.1093/emboj/19.19.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiRusso CC, Heimert TL, Metzger AK. Characterization of FadR, a global transcriptional regulator of Fatty scid metabolism in Escherichia coli. J. Bio. Chem. 1992;267:8685–8691. [PubMed] [Google Scholar]
- 36.DiRusso CC, Black PN, Weimar JD. molecular inroads into the regulation and metabolism of fatty acids, lesson from bacteria. Prog. Lipid Res. 1999;38:129–197. doi: 10.1016/s0163-7827(98)00022-8. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A. 1976;32:922–923. [Google Scholar]
- 38.Fliess A, Motro B, Unger R. Swaps in protein sequences. Proteins. 2002;48:377–387. doi: 10.1002/prot.10156. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Eisenberg D. 3D domain swapping: As domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rigali S, Derouaux A, Giannotta F, Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 2002;277:12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- 41.Gao YG, Yao M, Itou H, Zhou Y, Tanaka I. The structures of transcription factor CGL2947 from C. glutamicum in two crystal forms: a novel homodimer assembling and the implication for effector-binding mode. Protein Sci. 2007;16:1878–1886. doi: 10.1110/ps.072976907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorelik M, Lunin VV, Skarina T, Savchenko A. Structural characterization of GntR/HutC family signaling domain. Protein Sci. 2006;15:1506–1511. doi: 10.1110/ps.062146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee MH, Scherer M, Rigali S, Golden JW. a new member of the GntR family, has plasmid maintenance functions in Anabaena sp. strain PCC 7120. J. Bacteriol. 2003;185:4315–4325. doi: 10.1128/JB.185.15.4315-4325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franco IS, Mota LJ, Soares CM, de Sa-Nogueira I. Functional domains of the Bacillus subtilis transcription factor AraR and identification of amino acids important for nucleoprotein complex assembly and ligand binding. J. Bacteriol. 2006;188:3024–3036. doi: 10.1128/JB.188.8.3024-3036.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Aalten DM, DiRusso CC, Knudsen J. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 2001;20:2041–2050. doi: 10.1093/emboj/20.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Heath RJ, Li Z, Rock CO, White SW. The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J. Biol. Chem. 2001;276:7373–17379. doi: 10.1074/jbc.M100195200. [DOI] [PubMed] [Google Scholar]
- 47.Huffman JL, Brennan RG. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 48.Georgi T, Engels V, Wendisch VFA. Regulation of L-lactate utilization by the FadR-type regulator LldR of Corynebacterium glutamicum. J. Bacteriol. 2008;190:963–971. doi: 10.1128/JB.01147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong JM, Taylor JS, Latour DJ, Iuchi S, Lin ECC. Three overlapping lct gens involved in L-lactate utilization by Escherichia coli. J. Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iuchi S, Lin ECC. a global regulatory gene in Escherichia coli mediating repressor of enzymes in aerobic pathways. Proc. Natl Acad. Sci. USA. 1998;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tangeny M, Mitchan WJ. Analysis of a catabolic operon for sucrose transport and metabolism in Clostridium acetobutylicum ATCC 824. J. Mol. Microbiol Biotechnol. 2000;2:71–80. [PubMed] [Google Scholar]