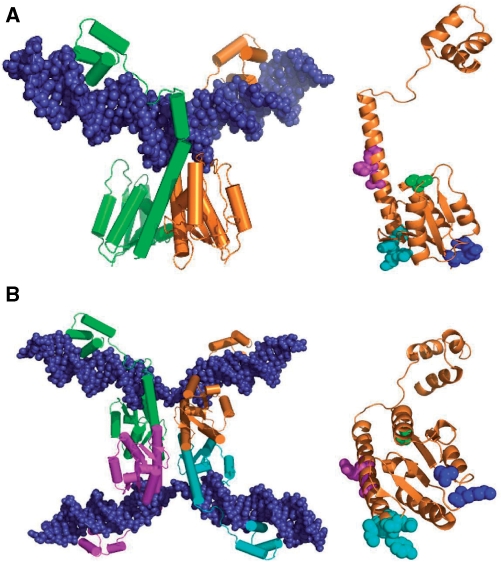
Figure 2.
Resolvase structures and activating mutations. (A) Crystal structure of a wild-type γδ resolvase dimer bound to site I [1GDT; (11)]. On the right, residues relevant to the experiments presented here are shown in spacefill on the orange resolvase subunit (in the same orientation); S10 in green; R2 and E56 in blue; G101, E102, M103 and K105 in cyan; A117, R121 and E124 in magenta. The hydroxyl group of S10 is the catalytic nucleophile in recombination. The sidechains of R2 and E56 contribute to the 2–3′ interface (see Introduction section). The other highlighted residues are the sites of activating mutations of Tn3 resolvase. The γδ resolvase residues E102 and K105 correspond to D102 and Q105, respectively, in Tn3 resolvase. (B) Crystal structure of a synaptic tetramer of γδ resolvase with two cleaved site Is [1ZR4; (16)], and on the right the orange resolvase subunit in the same orientation, showing residues in spacefill as in (A), except that the following residues are mutant; A2, K56, S101, Y102, I103, Q124. The images were created with PyMol.