Figure 6. Proposed model of RNA synthesis by VP1.
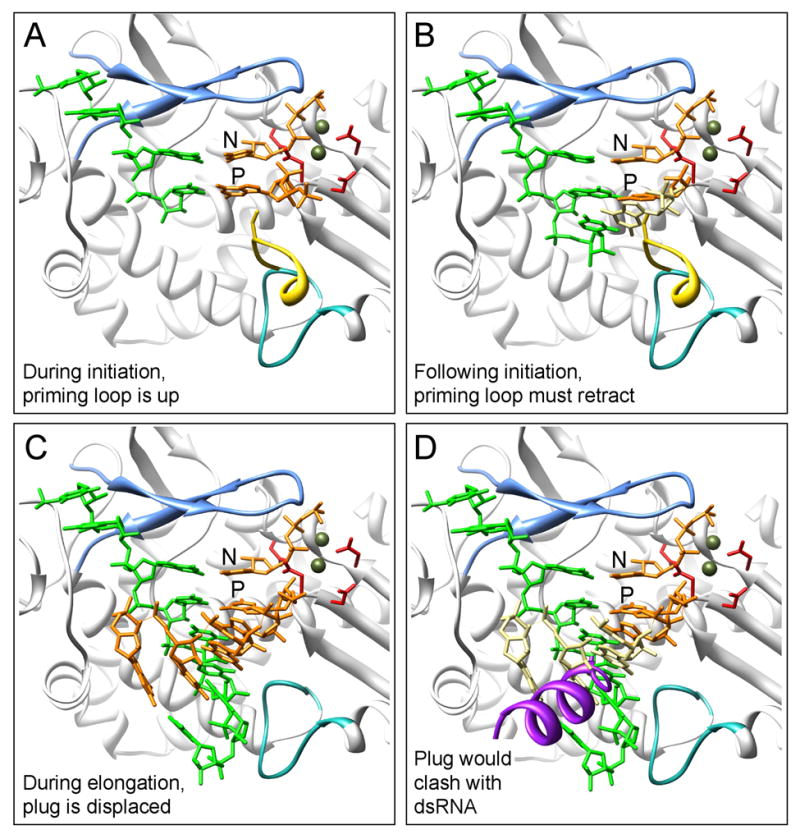
Superimposed on the VP1 structure are elements from the reovirus λ3 (A) initiation complex, PDB-1MWH (B) short elongation complex, PDB-1N38 and (C-D) long elongation complex, PDB-1N35. The reovirus template is shown in green, the incoming nucleotides, in orange, the Mg2+ ions, in olive, and the extended λ3 priming loop, in yellow. The priming (P) and incoming (N) nucleotide positions are indicated in panel (A). Important VP1 structural elements are shown, including: motif F (light blue), the priming loop (turquoise), and the catalytic aspartates (red). (A) During initiation of RNA synthesis, an extended priming loop is proposed to support the priming nucleotide, as seen for λ3. (B) Following initiation, translocation of the dinucleotide requires retraction of the priming loop to prevent a clash between the elongating product (beige) and the extended priming loop. (C) During elongation, a dsRNA product would emerge through the large exit tunnel when the VP1 C-terminal plug is assumed to be displaced. (D) An inserted C-terminal plug (purple) would conflict with passage of the dsRNA product (affected nucleotides in beige), requiring either repositioning of the plug during replication and/or redirection of the +RNA product during transcription.
