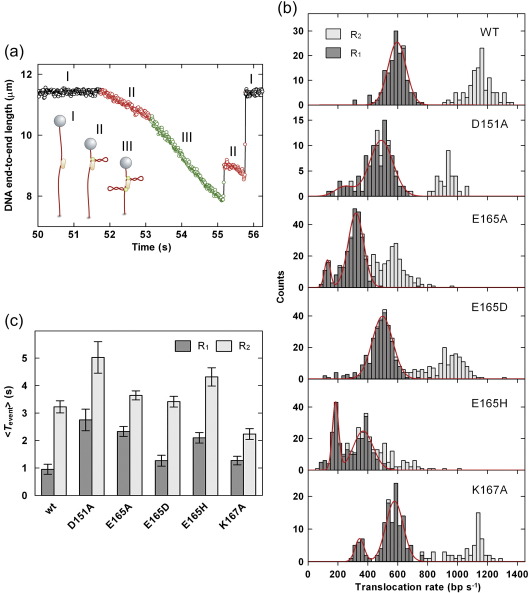
Fig. 3.
Single-molecule measurement of DNA translocation for WT and mutant HsdR subunits by magnetic tweezers. For all experiments, HsdR was at 160 nM, MTase was at 20 nM and F = 1.5 pN. (a) Sample single-molecule profile showing DNA length as a function of time. The inset shows a cartoon illustrating the principle of the assay, with DNA shown in red, the EcoR124I recognition site shown as a white box, the MTase shown as a light orange ovoid, the HsdR subunits shown as green circles and the magnetic bead shown in gray. See the main text for further details. The WT profile shows a period of inactivity, initiation of translocation of one HsdR subunit, initiation of translocation of a second HsdR subunit so that both motors are running simultaneously, dissociation of one subunit to leave a single translocating HsdR and, finally, dissociation of the remaining HsdR. (b) Histograms showing the translocation rates obtained from individual events binned in 25 s windows. Events are scored as either a single translocating HsdR (R1) or the sum of two translocating HsdR subunits (R2) (see the main text and Materials and Methods). Red lines are the fits of the R1 data to a single Gaussian (WT, E165D) or double Gaussian (D151A, E165A, E165H, K167A). Mean translocation rates for R1 population (from the single Gaussian fits) or both R1 and R1,slow populations (from the double Gaussian fits) are given in Table 2. (c) Histogram showing the average time for a translocation event (Tevent) when either one HsdR subunit (R1) or two HsdR subunits (R2) are translocating. Error bars represent the standard deviation of the mean. For HsdR subunits with bimodal R1 populations, Tevent represents the average of both subpopulations.