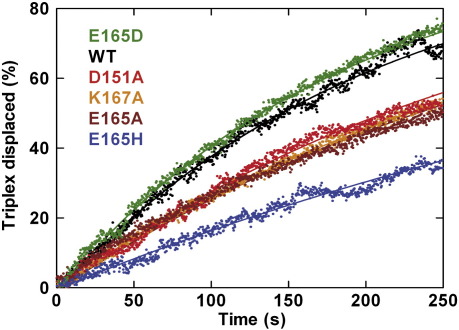
Fig. 6.
Bulk-solution measurement of DNA translocation and HsdR turnover by WT and mutant HsdR subunits using triplex displacement.19 Triplex displacement profiles were derived from stopped-flow experiments at a subsaturating HsdR concentration. Reactions were initiated by mixing preincubated triplex DNA (2093 bp between the EcoR124I and triplex binding sites), MTase and HsdR with an equal volume of reaction buffer plus ATP. The final solution contains 40 nM MTase, 1 nM HsdR, 5 nM DNA, 2.5 nM triplex and 4 mM ATP. Note that only 25% of the translocation events can lead to triplex displacement since there are 10 nM HsdR binding sites available (2 per DNA-bound MTase) but only 2.5 nM TFO bound.19 Experimental data are shown as scatter points, and continuous lines are fits to a single exponential.