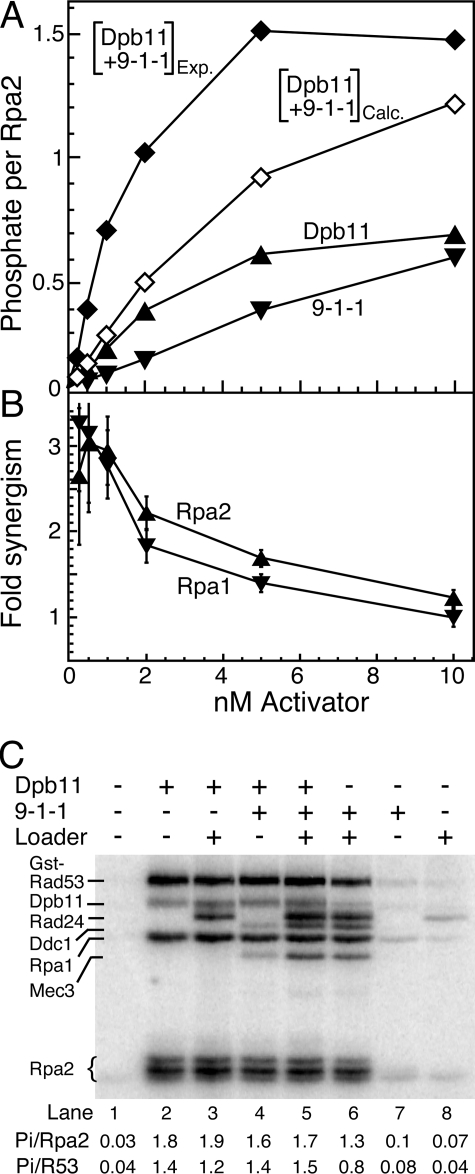
FIGURE 5.
Synergism between Dpb11 and 9-1-1. A, standard phosphorylation assays contained 1 nm deca-primed ssSK2 DNA, 150 nm RPA, 10 nm Rad24-RFC, and 5 nm Mec1, with the indicated concentrations of activator, either Dpb11, 9-1-1, or Dpb11 plus 9-1-1, at equimolar concentration. Assays were analyzed by 10% SDS-PAGE, and phosphorylation of Rpa2 was quantified and plotted. Open diamonds, calculated activity obtained by adding together the Dpb11 and 9-1-1-stimulated activities; closed diamonds, actual measured activity from the equimolar mixture of Dpb11 and 9-1-1. B, synergism, defined as the ratio of Mec1 stimulation activity by Dpb11 + 9-1-1 measured over that calculated from the individual assays (Dpb11 + 9-1-1)Exp/(Dpb11 + 9-1-1)Calc for Rpa2 phosphorylation from the data in A, and analogously for Rpa1 phosphorylation. C, assays as in A with 30 nm Dpb11, 30 nm 9-1-1, and 30 nm Rad24-RFC, as indicated. Quantification of Rpa2 and GST-Rad53-kd (R53) is given below the figure. In this assay GST-Rad53-kd was used rather than Rad53-kd to obtain a clear separation from other phosphorylated species. The GST tag does not alter the rate of phosphorylation (9).