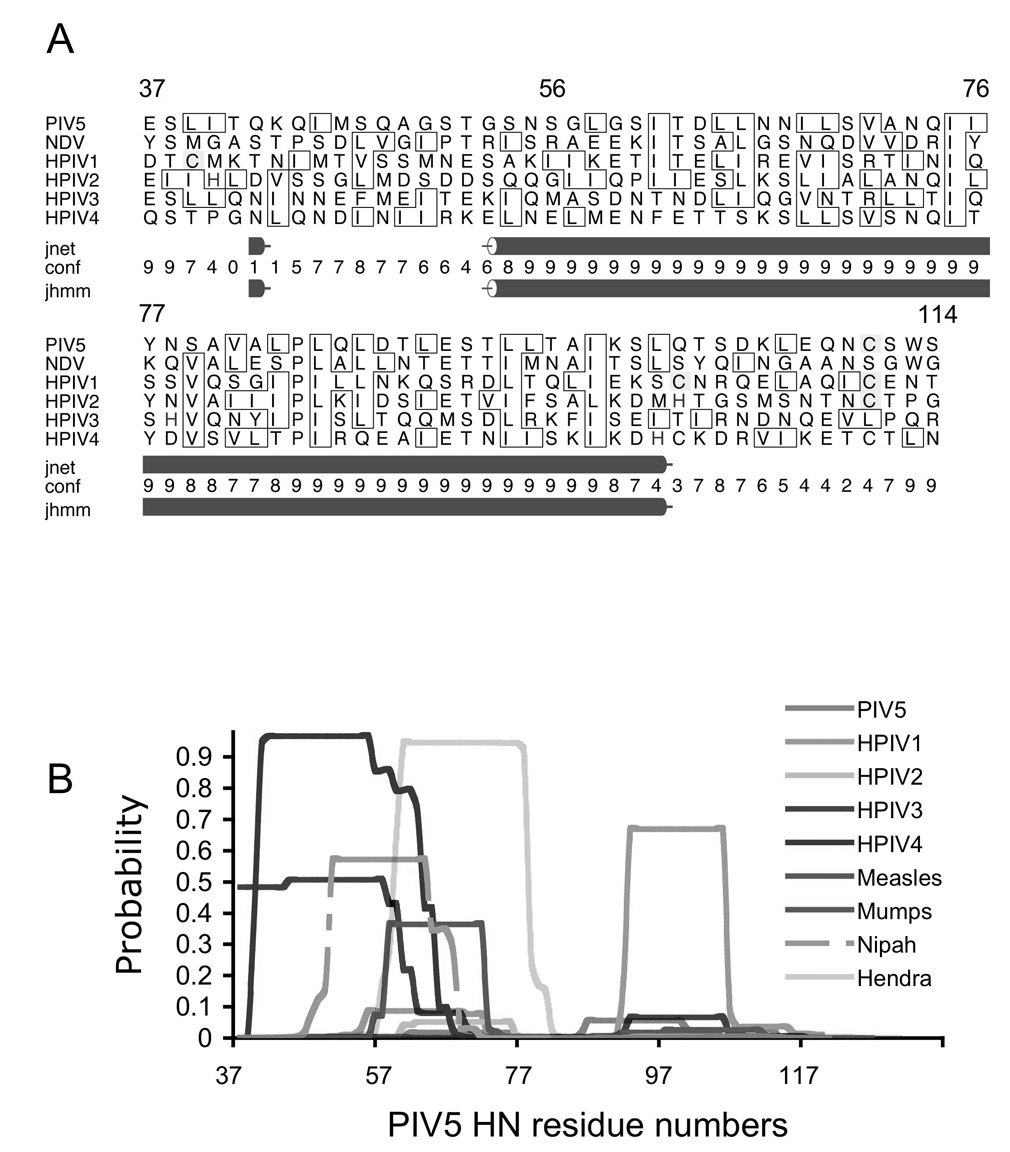
Figure 2. Sequence analysis of paramyxovirus attachment protein stalk regions.
(A) Secondary structure prediction based on multiple sequence analysis generated by the online Jpred server (http://www.compbio.dundee.ac.uk/~www-jpred/; University of Dundee, UK). The rows below correspond to the following: jnet: Jnet prediction; conf: an estimation of prediction accuracy on a scale 0–9; jhmm: Jnet hmm profile prediction(Cuff and Barton, 2000; Cuff et al., 1998). PIV5 HN residue numbering is indicated above the sequence alignment. (B) Predicted coiled coil propensity for stalk regions using the program COILS (http://ch.embnet.org/software/COILS_form.html; (Lupas, Van Dyke, and Stock, 1991)). The PIV5 HN stalk region exhibits a very low coil propensity, whereas other paramyxovirus attachment proteins have highly variable predictions, both in the calculated coiled coil probability and the specific residues implicated in coiled coil formation.