Abstract
Determination of the number of founders responsible for the establishment of invasive populations is important for developing biologically based management practices, predicting the invasive potential of species, and making inferences about ecological and evolutionary processes. The fire ant Solenopsis invicta is a major invasive pest insect first introduced into the USA from its native South American range in the mid-1930s. We use data from diverse genetic markers surveyed in the source population and the USA to estimate the number of founders of this introduced population. Data from different classes of nuclear markers (microsatellites, allozymes, sex-determination locus) and mitochondrial DNA are largely congruent in suggesting that 9–20 unrelated mated queens comprised the initial founder group to colonize the USA at Mobile, Alabama. Estimates of founder group size based on expanded samples from throughout the southern USA were marginally higher than this, consistent with the hypothesis of one or more secondary introductions of the ant into the USA. The rapid spread and massive population build-up of introduced S. invicta occurred despite the loss of substantial genetic variation associated with the relatively small invasive propagule size, a pattern especially surprising in light of the substantial genetic load imposed by the loss of variation at the sex-determination locus.
Keywords: biological invasion, colonization, invasive species, founder number, population bottleneck, propagule size
1. Introduction
Species of plants and animals that have successfully invaded areas beyond their natural ranges are of increasing concern to biologists, conservationists and public policy makers. Such invasive species often pose problems for the well-being of natural communities and ecosystems, agricultural production and the public health (Pimentel et al. 2000; Schlaepfer et al. 2005; Pauchard & Shea 2006; Gilchrist & Lee 2007), and the pace of biological invasions can only be expected to grow in the face of increased global trade and travel. Among the most enduring concerns about invasive species are as follows. Where and when did the colonists arrive in their new range? From where did the colonists originate? By what means were the colonists transported from their native range to the new area? and How many founders constituted the original colonizing population? In practical terms, answers to such questions are important for developing effective management practices based on the biology of the invasive species, as well as for predicting the invasive potential of other species (Sakai et al. 2001; Roderick & Navajas 2003; Strayer et al. 2006; Kang et al. 2007; Lavergne & Molofsky 2007; Miura 2007). From a scientific standpoint, the answers can inform a wide variety of topics of fundamental biological and conservation relevance, including the demographic responses of wild populations to population collapses, the resilience of ecosystems to perturbation, the evolution of novel traits in new selective environments and the population genetic consequences of bottlenecks (Carroll et al. 2005; Hoffmeister et al. 2005; Sax et al. 2005; Callaway & Maron 2006; Gilchrist & Lee 2007; Pascual et al. 2007; Tanaka et al. 2007; Zayed et al. 2007).
The red imported fire ant, Solenopsis invicta, is an introduced pest insect in the USA and other parts of the world which is considered by the World Conservation Union to be among the top 100 worst invasive alien species (IUCN 2000). This ant is native to South America, where it inhabits a vast range covering much of the centre of the continent. Owing to its status as a major pest throughout much of the southern part of the USA, an enormous amount of research has been conducted on the basic biology of the species over the past 40 years, making it one of the better known invasive organisms (e.g. Tschinkel 1993, 2006; Ross & Keller 1995; Gotzek & Ross 2007). This volume of information has in turn contributed to the emergence of S. invicta as a model for basic evolutionary and ecological research.
Improved knowledge of the reproductive biology, population demography, genetics and invasion history of S. invicta now puts us in an excellent position to make well-informed estimates of the size of the founder population responsible for the establishment of the species in the USA. It is clear that the ant first appeared near the port of Mobile, Alabama, sometime in the mid-1930s following maritime transport in unknown cargo (reviewed in Tschinkel 2006). Because fire ant workers are sterile, the invasive propagules presumably comprised newly mated queens blown onto the cargo during their mating flights or fragments of colonies containing reproductive queens as well as a worker force. It is also clear that the newly established population expanded rapidly in the decades following the initial founding. Recent population genetic surveys throughout the introduced range substantiate the close genetic resemblance of ants collected near Mobile to a hypothetical reconstructed ancestral population, but also raise the possibility of a secondary introduction into the USA at a location considerably west of Mobile (Shoemaker et al. 2006). Crucial to the task of estimating the invasive propagule size, the identity of the native S. invicta population from which all the US colonists originated is now reasonably well established (Caldera et al. 2008); this information is especially relevant in light of the pronounced regional differences in genetic diversity in the native range (Ross et al. 2007). Both the putative source and invasive populations have been well characterized genetically by assaying variation at a diverse assemblage of markers, including markers of both the nuclear and mitochondrial genomes (Ross et al. 1993, 1997; Shoemaker et al. 2006; Ross et al. 2007). These genetic data, along with relevant natural history information, are incorporated here in traditional and new analytical approaches to infer the make-up of the original founder population of S. invicta in the USA. Among other purposes, this information can be used to postulate probable scenarios for the introduction of this and other ants with similar habits into other parts of the world, as well as to help provide a general foundation for understanding how the loss of genetic diversity often associated with founder events may influence the evolution of invasive populations.
2. Material and methods
(a) Samples and genetic data
Most of the S. invicta samples used in this study have been described in detail in previous papers (Shoemaker et al. 2006; Ross et al. 2007). These samples were obtained from two localities in the native South American range (Formosa and Corrientes, Argentina; see Ross et al. 2007) and six localities distributed across the introduced US range (Texas, west Louisiana, east Louisiana, Mississippi, Georgia and Florida; see Shoemaker et al. 2006). The northern Argentina population from Formosa was previously determined with high confidence to be the source of the original US colonists based on combined nuclear and mtDNA data, although some potential contribution from the neighbouring (160 km distant) Corrientes population was suggested as well by the mtDNA data (multiple lines of evidence effectively ruled out all the sampled native populations other than these two as potential sources of the US colonists; Caldera et al. 2008). Thus, we considered either the Formosa samples alone or the combined Formosa and Corrientes samples as sources of the US founder population in our analyses. Among the six US localities, we consider the Mississippi population as the exemplar introduced population because it is located near (50 km from) the primary site of the entry of S. invicta into the USA (Mobile, Alabama) and because it appears to have diverged little genetically from the hypothetical founder population there (Shoemaker et al. 2006). We used either the Mississippi samples alone or the entire set of the US samples to represent the colonizing population in different analyses; inclusion of the latter expanded set of samples is particularly important in evaluating the possible impact of a secondary introduction of S. invicta into the USA following the primary introduction into Mobile (see Shoemaker et al. 2006). Nests of both social forms (monogyne (single queen per nest) and polygyne (multiple queens per nest)) were sampled at each of the eight principal sampling sites in the two ranges; data from the two nest types at each site were combined owing to the high overall genetic similarity of the forms where they occur in sympatry (Shoemaker et al. 2006; Ross et al. 2007). The total numbers of the sampled colonies are 37, 89, 163 and 382 for the mtDNA, and 70, 158, 131 and 1038 for the standard nuclear loci, for the Formosa, combined Argentina, Mississippi and combined US datasets, respectively.
Most of the data from standard genetic markers that we analyse in this study originally were described by Ross et al. (2007; native populations), Shoemaker et al. (2006; standard nuclear data in US populations), Caldera et al. (2008; mtDNA data in major US populations) and Ross et al. (1997; Est-4 in native populations). These standard data comprise individual genotypes scored at 14 nuclear markers (7 allozyme and 7 microsatellite loci) and individual sequences determined for a 920 bp fragment of mtDNA. New mtDNA sequence data were generated for this study for 118 colonies from all of the major US populations other than west Louisiana (methods in Ahrens et al. 2005). New mtDNA data were generated as well from two additional US populations (16 colonies from Demopolis, Alabama, and 19 colonies from Vaughan, Mississippi). (All mtDNA sequences newly generated for this study have been deposited in GenBank under accession numbers EU352605–EU352610 and EU373818–EU373820.) The numbers of variants (allele or haplotype richness) observed for the different markers in each focal population or group of populations are reported in table 1. With the possible exception of the esterase genes Est-2 and Est-4 (Ross & Shoemaker 2005), linkage disequilibrium between pairs of the nuclear genes is unimportant in native S. invicta (Ross et al. 2007); thus, these can largely be regarded as independent markers of population history. Because fire ant colonies are typically composed of related nest-mates, only a single individual (genotype or haplotype) per nest was used in our analyses of the standard markers in order to ensure unbiased estimates of population genetic diversity.
Table 1.
Numbers of variants observed for genetic markers used in this study. (Argentina samples are pooled from Formosa and Corrientes; US samples are pooled from six sites located across the colonized range. The estimate of the number of sex-determination alleles for the native range is from the polygyne social form in Corrientes, while that for the colonized range is from the monogyne social form in Georgia (Ross et al. 1993). The number of mtDNA haplotypes in the USA includes the new sequences generated for this study.)
| marker | native range | colonized range | ||
|---|---|---|---|---|
| Formosa | Argentina | Mississippi | USA | |
| allozymes | ||||
| Aat-2 | 2 | 3 | 2 | 2 |
| Acoh-1 | 5 | 6 | 2 | 2 |
| Acoh-5 | 3 | 3 | 2 | 2 |
| Est-2 | 3 | 7 | 1 | 1 |
| Est-4 | 2 | 4 | 2 | 2 |
| G3pdh-1 | 2 | 2 | 2 | 2 |
| Pgm-1 | 4 | 6 | 2 | 2 |
| microsatellites | ||||
| Sol-6 | 12 | 17 | 4 | 6 |
| Sol-11 | 14 | 15 | 5 | 8 |
| Sol-18 | 5 | 7 | 3 | 3 |
| Sol-20 | 14 | 19 | 3 | 12 |
| Sol-42 | 22 | 26 | 9 | 14 |
| Sol-49 | 17 | 18 | 6 | 11 |
| Sol-55 | 16 | 16 | 7 | 8 |
| sex-determination locus | — | 86 | — | 12 |
| mtDNA | 18 | 37 | 6 | 9 |
Data from a novel non-standard nuclear marker also were employed in this study to provide independent estimates of the numbers of founders. A single major gene is hypothesized to regulate sex determination in S. invicta by means of allelic complementation (Ross & Fletcher 1985a; Ross et al. 1993; see also Beye 2004; Zayed et al. 2007; Heimpel & de Boer 2008). In such systems, the effective numbers of alleles at the sex-determination locus can be estimated indirectly from the population-wide frequencies of ‘matched matings’, those matings that yield atypical diploid males (Adams et al. 1977; Cook & Crozier 1995). In S. invicta, frequencies of occurrence of such matings have been estimated for the native Corrientes population (based on 95 queens from 15 polygyne nests) and for several invasive US populations, the most comprehensive estimate being that for a monogyne population in Georgia (based on 648 queens derived, presumably, from as many nests; Ross et al. 1993). Owing to strong negative frequency-dependent selection acting on such systems, the actual number of segregating alleles approaches the effective number, with each of the K alleles expected to occur at a frequency of 1/K (Yokoyama & Nei 1979; Cho et al. 2006). We used this information to infer the numbers of sex-determination alleles, their population frequencies and their counts within samples in Corrientes and Georgia. We note that the number of sex-determination alleles in the native Formosa population is likely to be very similar to that in neighbouring Corrientes, given the similar proportions of diploid males observed at the two sites (Ross et al. 1993). Moreover, the number estimated for Georgia is likely to be representative of all invasive S. invicta in the USA, given the similar proportions of diploid males or matched matings detected at five sites from Texas to Florida (Ross et al. 1993), including the Mississippi site sampled for the other markers.
(b) Estimation of numbers of founder queens
The simplest method to estimate the number of founders of the US S. invicta population was to determine the minimum number of mated queens required to explain the observed allele and haplotype richness. For these and other analyses, we assumed that each founder queen had mated with a single (haploid) male, an assumption supported by the studies of native as well as introduced S. invicta (Ross & Fletcher 1985b; Ross et al. 1993, 1997). We further assumed that mtDNA heteroplasmy is rare or non-existent in S. invicta (multiple peaks at single positions have never been observed in the mtDNA sequence traces obtained from the large samples of individuals from both the native and introduced ranges; e.g. Ahrens et al. 2005; Caldera et al. 2008; this study). Therefore, each founder queen could carry at most three distinct nuclear alleles per locus and one mtDNA haplotype.
A second method to estimate founder numbers used computer simulations to model the effects of the founding event on mtDNA haplotype richness (e.g. Murray-McIntosh et al. 1998; Ross et al. 2003; Zayed et al. 2007; Jackson et al. 2008). The simulations involved random sampling, with replacement, of different numbers of haplotypes (mated founder queens) from the lists of haplotypes and their counts in the assumed source populations. Sampling for each source population and number of founders was repeated 1000 times to generate distributions depicting the probabilities that different numbers of haplotypes were established by various numbers of founders. The estimates obtained by this simulation method represent the genetically effective numbers of founders rather than the actual numbers, because they do not account for the loss of haplotypes through drift subsequent to the initial founder event.
For these mtDNA simulations, corrected estimates of the numbers of haplotypes that occur in the native populations were obtained by two methods. The first involved using a modified rarefaction analysis to generate a haplotype discovery curve (Jackson et al. 2008). The original haplotype data were subsampled 10 000 times for each of 20 different sample sizes and a one-phase exponential association function was fitted to the 20 mean values by means of nonlinear regression using the program GraphPad Prism (this particular function was chosen based on the goodness of fit and shape of the curve for interpolation). Values of Ymax in the function represent the haplotype richness at the asymptote. The second method of obtaining a corrected estimate of haplotype richness was calculation of the non-parametric abundance-based Chao1 estimator (Chao 1984). Undiscovered haplotypes assumed to exist based on the corrected estimates were represented as single haplotypes in the modified native haplotype lists (such haplotypes presumably were sufficiently rare to have been missed in our samples). Also, one US haplotype not recovered from any native population but which differs from a Formosa haplotype by only a single substitution (Caldera et al. 2008) was assumed to be present in all source populations, where it was represented on the list by a single individual. Finally, one US haplotype recovered from Corrientes but not Formosa was assumed to occur in the latter population as well when this was specified as the source (again, represented by a single individual).
Our corrected estimates of the haplotype richness in the native Formosa, Argentina population are 25.1 (rarefaction analysis) or 27.2 (Chao1 estimator), compared with the 18 haplotypes actually recovered there. Corrected estimates of the richness in all the Argentina samples are 52.6 or 54.4, respectively, compared with the 37 haplotypes recovered. As expected, given the large sampling effort and low haplotype diversity in the USA, application of the Chao1 estimator yielded effectively no increase in the corrected over the observed haplotype richness for both Mississippi and the combined US samples.
A final method for estimating the number of mated founder queens, in this case using the nuclear data, involved application of a likelihood-based estimator originally developed to estimate effective population size (Ne) based on coalescent theory (Berthier et al. 2002; Anderson & Slatkin 2007). The method considers the ancestry of single loci scored in known source and colonizing populations as representing the genealogy of the colonizing population, assuming that the gene genealogies follow a neutral coalescent process for a given Ne and a given number of generations. The likelihood that the samples from the invasive range are descended from a ancestral lineages (chromosomes) extant T generations previously is calculated approximately via a Monte Carlo importance sampling algorithm (Anderson 2005; Anderson & Slatkin 2007). We used the program CoNe (Anderson 2005) to evaluate the likelihoods of different values of a based on 5000 Monte Carlo replicates in each simulation for the different source and colonizing populations.
Approximately 60 years separate the first arrival of S. invicta in Mobile and the collection of our US samples. We assume that almost all of the early expansion, as well as much of the subsequent population build-up, in the invasive range is attributable to the monogyne social form (Shoemaker et al. 2006). Colonies of this form generally begin producing new sexuals at 3 years of age, and continue producing these in large numbers through the 7- to 8-year life of the colony (queen), with larger (older) colonies producing disproportionately higher numbers of sexuals (Tschinkel 2006). Colonies in suitable unoccupied habitat can be expected to have higher reproductive success, and shorter times to first successful reproduction, than colonies in fully occupied environments; thus, average generation time was probably shorter early in the invasion process than more recently. Given these complexities as to what constitutes generation length in fire ants, we set the number of generations that have passed between founding and sampling of the US S. invicta population at either 10 or 20 in the different CoNe simulations.
Estimation of the number of founder lineages or chromosomes remaining at the time of sampling (a) via the methods in CoNe assumes that the samples for each locus are independent (no linkage disequilibrium), that mutation at the marker genes was negligible in the colonizing population and that drift in the source population was negligible between the founder event and sampling compared with the genetic changes in the colonizing population (Berthier et al. 2002; Anderson & Slatkin 2007); these assumptions have been confirmed for S. invicta (unimportance of disequilibrium) or are likely to be true, given our knowledge of this species and its introduction. Berthier et al. (2002) showed with simulated datasets that the total number of alleles is the most important factor affecting the precision of estimates of a, but that the numbers of loci and samples also are important. Thus, our dataset, which features large sample sizes and a number of highly polymorphic loci, may be expected to yield quite reliable estimates of a. We used CoNe to generate likelihood curves for each locus and for the combined nuclear data, as well as 95% confidence intervals around the overall maximum-likelihood estimates of a (Anderson 2005). Estimates of a were divided by three to deduce the number of unrelated mated founder queens that left some genetic legacy (the genetically effective founder number).
(c) Simulations of founder population growth and loss of genetic diversity
Our estimates of founder group size derived from the mtDNA founding simulations and nuclear gene CoNe simulations are expected to closely reflect the actual number of founders only if early colonizing population growth rates were high and the number of founders was relatively low (Knowles et al. 1999; Anderson & Slatkin 2007). In other circumstances, these approaches provide underestimates of the numbers of founders because, in accounting for the variants that presently exist in the US range, they do not distinguish between the primary loss of diversity at initial founding and subsequent secondary losses of diversity caused by drift in a finite population. Said differently, only a subset of the founders may be expected to have their genetic lineages represented in the extant invasive population, in which case a is an estimate of the genetically effective rather than actual number of founder lineages. Our results indicate that S. invicta may well have colonized the USA with 10 or fewer mated queens (30 chromosomes), and knowledge of the reproductive biology and early spread of this ant suggests that the invasive propagules may have experienced early intrinsic (exponential) population growth rates in excess of one (Morrill 1974; Vargo & Fletcher 1987; Tschinkel 2006); these founder numbers and growth rates are considered threshold values for the genetically effective and actual numbers of founder lineages to be similar (Anderson & Slatkin 2007). Nonetheless, it is prudent to examine how specific features assumed for the invasion and demographic history of the founder population may affect the relationship between the effective and actual numbers of founders. Therefore, we conducted computer simulations to study the general patterns of the secondary loss of diversity following the initial founding event under different assumptions of the size and genetic diversity of the founder group and of early population growth rate (see also Noor et al. 2000; Ross et al. 2003; Ficetola et al. 2008).
These simulations assumed that the founder population consisted of 9, 12 or 20 singly mated unrelated queens, numbers chosen because they generally bracket our estimates of genetically effective numbers of ancestral queens inferred by the various methods. Simulations were carried out for the most polymorphic markers (six microsatellite loci, the sex locus and the mtDNA; table 1). The numbers of unique haplotypes or alleles assigned to the founder group ranged from the number currently found in the USA to two or three variants in excess of this number. For simulations involving the sex locus, allele counts were equalized as much as possible within the founder group (i.e. variance in allele frequencies was minimized), in accordance with the expected homogeneous frequencies of such alleles in the source population. Counts of the mtDNA and microsatellite variants were set to reflect the observed frequencies of the most common variants in Formosa, Argentina, the likely sole or major source of the US colonists (Caldera et al. 2008).
Rates of population increase (r) for these simulations were set at five values varying from 0.2 to 1.2 (20–120% increase in colony (reproductive queen) number every generation). The variants that populated each simulated generation were selected at random (with replacement) from the previous generation, and the simulations were carried out until carrying capacity was reached (1000 queens for the mtDNA and 2000 adult sexuals for the nuclear markers). Depending on the values of r and initial founder size, from 6 to 26 generations were required to reach carrying capacity, at which point we assume no further erosion of diversity would have occurred. The simulations for the nuclear markers additionally incorporated Hardy–Weinberg genotype proportions and random mating with respect to genotype (see Ross et al. (1997, 1999, 2007) and Shoemaker et al. (2006) for evidence that these assumptions are valid for S. invicta in the native and US ranges), as well as equal allele frequencies in the sexes and equal numerical sex ratios throughout the simulations. Probabilities of maintenance of the allele and haplotype richness now observed throughout the US range were estimated as the proportions of 1000 iterations yielding at least this number of extant variants once carrying capacity was reached.
3. Results and discussion
Examination of the allele and haplotype richness (number of variants) in invasive S. invicta offers the simplest means of determining the minimum number of mated founder queens responsible for the establishment of the species in the USA. The highest estimates for Mississippi and the total US population, six and nine, respectively, correspond to the numbers of mtDNA haplotypes recovered. Data inferred for the most polymorphic nuclear gene, the sex-determination locus, suggest a minimum of four founders for either population, while allele richness at the most polymorphic microsatellite (Sol-42) is consistent with a minimum of three and five founders for the respective populations. These are almost certainly underestimates of the numbers of founders, given that they are limited by the polymorphism of the markers and that they assume every founder queen bore a unique haplotype and a set of three unique alleles per nuclear gene different from every other founder.
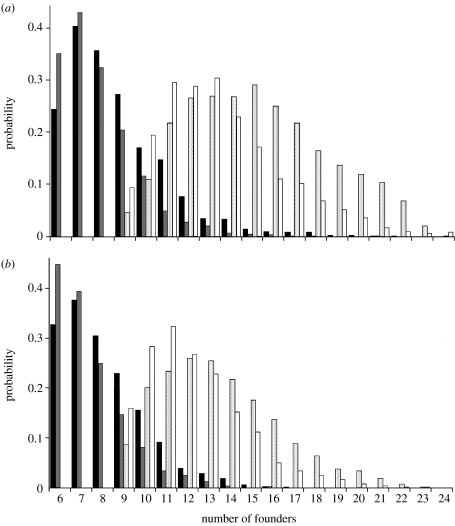
Results of computer simulations modelling the effects of founding on mtDNA haplotype richness are shown in figure 1. Given the haplotype data from the native range, the most likely number of founders bearing six haplotypes, the number currently found in Mississippi, is six or seven, depending on whether Formosa or the pooled Argentina samples are considered as the potential source and whether or not estimates of the native haplotype richness are corrected. In all cases, only founder group sizes from six to nine yield probabilities greater than 0.2 of carrying six unique haplotypes. The most likely number of founders bearing nine haplotypes, the number currently found throughout the USA, ranges from 10 to 17 (probabilities greater than 0.2). These probabilities probably underestimate the propagule size because additional losses of haplotypes may have occurred due to drift following the initial founder event; thus, they are best viewed as probabilities of the genetically effective numbers of founders.
Figure 1.
Simulation-derived probabilities of six or nine mtDNA haplotypes being represented in different-sized groups of founder queens of S. invicta in the USA. Six haplotypes presently occur in Mississippi, whereas nine occur throughout the US range. Sources of the founders are assumed to be (a) Formosa or (b) Argentina (pooled Formosa and Corrientes samples). Simulations were run using both the uncorrected (observed) and corrected estimates of haplotype richness in the source populations. Black bars, six haplotypes, uncorrected richness; grey bars, six haplotypes, corrected richness; stippled bars, nine haplotypes, uncorrected richness; white bars, nine haplotypes, corrected richness.
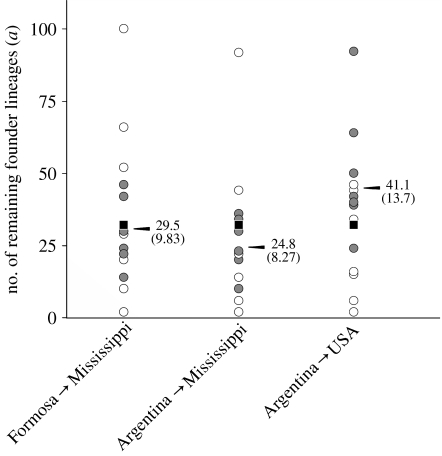
Likelihood curves for estimates of a, the number of remaining haploid founder lineages, obtained from coalescence-based CoNe simulations are shown for each nuclear gene for various colonization scenarios in figure S1 of the electronic supplementary material. Maximum-likelihood estimates of a derived from the combined nuclear data correspond to genetically effective founder group sizes that are remarkably similar to those obtained from the mtDNA founder simulations described above, when 10 generations between founding and sampling are assumed (figure 2). Thus, the nuclear simulations suggest a high likelihood of 8–10 founders of the Mississippi population, while the mtDNA simulations suggest a high probability of 6–9 founders for this single population (lower values are favoured in each case when the larger and more diverse source population is assumed). Also, the maximum-likelihood value of approximately 14 founders of the entire US population derived from the nuclear data is bracketed by the 10–15 queens representing the highest probabilities of founding this population based on the mtDNA simulations (assuming Argentina as the source in both cases). The maximum-likelihood estimates of founder number based on the nuclear simulations effectively double when the number of generations is assumed to be 20 rather than 10 (electronic supplementary material, figure S1), illustrating the sensitivity of the coalescence-based method to the assumed values of this variable. Nonetheless, the estimates are in all cases less than 30, and generally less than 20 founder queens. We note again that the results from the nuclear coalescence simulations may be underestimates of the actual founder numbers (they amount to the genetically effective founder numbers) because they do not account for any loss of lineages that occurred through drift following founding.
Figure 2.
Single-gene maximum-likelihood values for the number of founder lineages remaining in invasive S. invicta in the USA (a) distinguished by the class of nuclear marker. Values are derived from the coalescent simulation program CoNe by assuming 10 generations between initial colonization and sampling. Numbers at arrowheads indicate the overall maximum-likelihood values of a for each introduction scenario, with the corresponding numbers of unrelated mated founder queens shown in parentheses. Open circles, allozymes; filled circles, microsatellites; squares, sex locus.
The maximum-likelihood coalescence-based estimates of a from the 10-generation simulations are shown separately for each nuclear gene in figure 2. When Mississippi is designated as the colonizing population, there is no evident bias in the estimates by marker type, although the higher diversity microsatellites clearly display less variance than the lower diversity allozymes. When the entire US population is considered, the microsatellites generally yield higher estimates than the allozymes. The estimates derived from the sex locus fall roughly in the middle of the range of values from the traditional nuclear loci in all cases. This pattern would seem to validate this highly novel, indirectly assayed marker as being useful for inferring genetically effective founder numbers. Identical relative rankings of the single-gene a values were obtained in the 20-generation simulations.
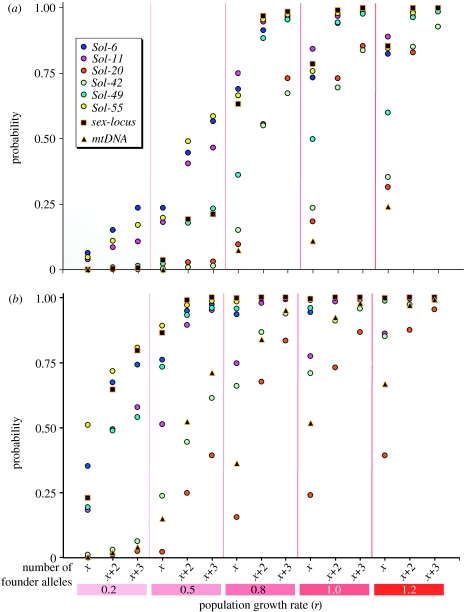
In the event that early founder population growth was high and the number of founders relatively low, estimates of the genetically effective founder numbers obtained by the mtDNA and nuclear coalescence-based simulations may closely approximate the actual propagule size (Knowles et al. 1999; Anderson & Slatkin 2007). Otherwise, only a subset of the original founder lineages is expected to be represented in our samples, with the other original lineages having been lost through drift. The relationship between the number of lineages at founding and the number surviving to the time of sampling depends on specific features of the demographic history of the colonizing population; hence, it is important to examine this relationship under different demographic scenarios (Anderson & Slatkin 2007). The results of our simulations in which founder group size and genetic composition, as well as population growth rate, were systematically varied to infer the probability of retaining the extant allele and haplotype richness in the USA are shown in figure 3 (for simplicity, only the extreme cases of 9 and 20 founders are shown). When population growth rates are modest (r≤0.5), probabilities of the extant numbers of variants being retained are often low (less than 0.5), even with 20 founders bearing three variants in excess of the extant number. However, with values of r of 0.8 or higher, the probabilities rise above 0.5 for all markers when two or three excess variants are assumed in as few as nine founders. Even when no excess variants are assumed, growth rates of r≥1 yield high probabilities that the extant numbers of variants are retained at most markers when the founder groups comprise 12 or 20 queens. Given the extraordinary reproductive potential of mature S. invicta colonies (Morrill 1974; Vargo & Fletcher 1987) and historical records of rapid population expansion following the initial founding (Tschinkel 2006), growth rates of r≥1 (100% or more increase in the number of colonies per generation) would seem biologically realistic for the early colonizing population (see also Ficetola et al. 2008). Thus, these simulations suggest that our estimates of the genetically effective number of mated founder queens may be quite close to the actual number, especially if the founder group bore a small number of variants in addition to those presently found in the USA at most markers. These results, based on empirically determined allele counts and frequencies from the native and introduced ranges of S. invicta, are congruent with the findings of Anderson & Slatkin (2007) based on simulated data.
Figure 3.
Probabilities of the numbers of genetic variants extant at the most polymorphic markers in S. invicta in the USA being retained, based on the simulations of different scenarios of the size and genetic diversity of the founder group and of early population growth rate (r). Probabilities are shown for invasive propagule sizes of (a) 9 or (b) 20 mated founder queens. For the number of alleles in the founder groups, x indicates the extant number, while x+2 and x+3 indicate two and three variants in excess of this number, respectively. Simulations could not be run for x+2 and x+3 mtDNA haplotypes with nine founders.
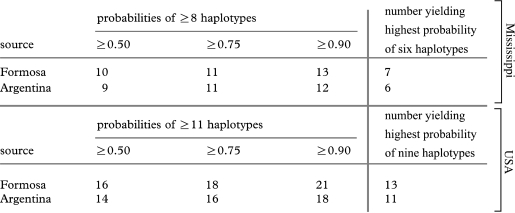
Given the effect of excess variants carried by the founder queens on the relationship between the genetically effective and actual founder numbers, it is worth revisiting the results of our mtDNA simulations of the impact of the founder group composition on haplotype richness. Considering Mississippi as the relevant introduced population, the probabilities of a propagule group from Formosa or the combined Argentina populations carrying two or more extra haplotypes become relatively high once 10 or 9 queens, respectively, comprise the group, a modest increase over the most probable numbers bearing six haplotypes (table 2). Considering the entire USA as the relevant introduced population, relatively high probabilities are attained once the numbers of queens reach 16 or 14, respectively, again a modest increase over the most likely numbers bearing 9 haplotypes (table 2). The revised figures shown in columns 2–4 of table 2 may be regarded as the best approximations of the actual numbers of mated founder queens based on the mtDNA data, whereas the unrevised figures in column 5 (obtained from figure 1) are best viewed as approximations of the genetically effective numbers.
Table 2.
Numbers of founder queens necessary to introduce extant numbers of mtDNA haplotypes or at least two in excess of extant numbers into the USA, based on simulations. (Data in the top part of the table are relevant to the Mississippi population, while data in the bottom part are relevant to the entire US population. Data in columns 2–4 concern the introduction of two or more mtDNA haplotypes in excess of the extant number, while those in column 5 concern the introduction of the extant number (figure 1). Simulations used native haplotype richness corrected for unobserved haplotypes; simulations using uncorrected richness yielded slightly higher numbers.)
 |
It is worth noting that estimates of either the actual or effective numbers of founders may be preferable, depending on the goals of an analysis. A sound estimate of the actual number may be desired, for instance, in evaluating the potential for the successful introduction of specific organisms via specific conveyance mechanisms, whereas an estimate of the genetically effective number would be more relevant to evaluating the loss of genetic diversity and its effects on the persistence and evolutionary potential of a colonizing population. For S. invicta, the actual numbers are appropriately regarded as genetically independent ‘queen equivalents’. For reproductive queens of the monogyne form or newly mated queens of either form that undertook dispersal flights, these equivalents should be the same as the actual number of individual founders. For polygyne reproductive queens, nest-mates of which are closely related in the native range (Ross et al. 1996), the equivalents may correspond more closely to the number of colonies from which the founder queens originated than to the number of individual queens.
The results of this study, based on large sample sizes, data from a number of highly informative genetic markers and relevant background biological information, provide the first robust estimates of the numbers of founder queens of S. invicta responsible for colonizing the USA. Based on the data from the Mississippi population, our analyses suggest that 9–20 unrelated mated queens are likely to have comprised the initial founder group to colonize the USA at Mobile, Alabama, in the mid-1930s. Estimates based on the complete set of pooled US samples range marginally higher, with perhaps 15–30 queens involved in the colonization of the entire introduced range. The consistent differences between the estimates of founder number derived from the Mississippi and the total US samples in all of our analyses can be taken as support for the hypothesis that one or more secondary introductions of the ant into the USA occurred (see also Shoemaker et al. 2006).
The congruence between the results based on the different classes of markers and different analytical approaches lends weight to the credibility of our estimates. Among the nuclear markers, the sex-determination locus stands out from the others with respect to the way that variability was assayed; allele counts and frequencies were inferred indirectly based on the proportions of queens producing atypical, diploid males, rather than directly by scoring individual genotypes. The mtDNA sequence data not only track a separate genome with a different mode of inheritance and effective population size than the other markers, but also these data were analysed using a completely different simulation method than the coalescent-based likelihood method used to analyse the nuclear data.
Our conclusion of a relatively small propagule size for invasive S. invicta in the USA is consistent with a pronounced bottleneck being the major cause of the severe reduction in the diversity of the invasive gene pool (table 1). This result is relevant to the general issue in invasion biology of whether significant losses of diversity typically accompany introductions, and if so, how these may affect the subsequent persistence and evolution of the invasive populations (Johnson & Starks 2004; Bossdorf et al. 2005; Sax et al. 2005; Hedrick et al. 2006; Gilchrist & Lee 2007; Lavergne & Molofsky 2007; Miura 2007; Zayed et al. 2007; Dlugosch & Parker 2008; Ugelvig et al. 2008). In invasive social insects, understanding specific features of founder events such as the numbers and origins of the colonists can be important for predicting an array of significant evolutionary responses, including alterations in colony social structure mediated by the loss of nest-mate recognition alleles (e.g. Giraud et al. 2002; Ross et al. 2003; Tsutsui et al. 2003) and shifts in the mating biology caused by the loss of sex-determination alleles (Ross et al. 1996). Indeed, in Hymenoptera with complementary sex-determination systems based on high allelic diversity (Ross & Fletcher 1985a; Cook & Crozier 1995; Heimpel & de Boer 2008), successful colonization by a small group of founders may seem an unlikely prospect altogether, because the enormous genetic load imposed on populations by substantial loss of sex alleles is expected in many circumstances to culminate in extinction (Zayed & Packer 2005; Hedrick et al. 2006). Yet, several examples of successful invasion by such species following severe population bottlenecks have been documented (Ruttner 1976; Ross & Fletcher 1985a; Schmid-Hempel et al. 2007; Zayed et al. 2007). In such cases, the emergence of new ecological opportunities coupled with release from the effects of natural enemies and competitors may more than offset the negative genetic consequences stemming from the same founder event (Ross & Fletcher 1986).
Acknowledgments
We thank Chris DeHeer, Laurent Keller, Mark Mescher, James Trager and Edward Vargo for their assistance in collecting samples. This study was supported by grants from the US Department of Agriculture NRICGP (nos. 036393-01 and 2006-35302-18001) and by the Georgia Agricultural Experiment Stations, University of Georgia. The use of trade, firm or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the US Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable.
Supplementary Material
Figure with likelihood values for ‘a’
Caption for electronic supplementary material figure S1
References
- Adams J, Rothman E.D, Kerr W.E, Paulino Z.L. Estimation of the number of sex alleles and queen matings from diploid male frequencies in a population of Apis mellifera. Genetics. 1977;86:583–596. doi: 10.1093/genetics/86.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens M.E, Ross K.G, Shoemaker D.D. Phylogeographic structure of the fire ant Solenopsis invicta in its native South American range: roles of natural barriers and habitat connectivity. Evolution. 2005;59:1733–1743. doi:10.1554/05-067.1 [PubMed] [Google Scholar]
- Anderson E.C. An efficient Monte Carlo method for estimating Ne from temporally spaced samples using a coalescent-based likelihood. Genetics. 2005;170:955–967. doi: 10.1534/genetics.104.038349. doi:10.1534/genetics.104.038349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.C, Slatkin M. Estimation of the number of individuals founding colonized populations. Evolution. 2007;61:972–983. doi: 10.1111/j.1558-5646.2007.00080.x. doi:10.1111/j.1558-5646.2007.00080.x [DOI] [PubMed] [Google Scholar]
- Berthier P, Beaumont M.A, Cornuet J.M, Luikart G. Likelihood-based estimation of the effective population size using temporal changes in allele frequencies: a genealogical approach. Genetics. 2002;160:741–751. doi: 10.1093/genetics/160.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beye M. The dice of fate: the csd gene and how its allelic composition regulates sexual development in the honey bee, Apis mellifera. Bioessays. 2004;26:1131–1139. doi: 10.1002/bies.20098. doi:10.1002/bies.20098 [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers W.E, Siemann E, Prati D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144:1–11. doi: 10.1007/s00442-005-0070-z. doi:10.1007/s00442-005-0070-z [DOI] [PubMed] [Google Scholar]
- Caldera, E. J., Ross, K. G., DeHeer, C. J. & Shoemaker, D. D. 2008 Putative native source of the invasive fire ant Solenopsis invicta in the USA. Biol. Invasions (doi:10.1007/s10530-008-9219-0)
- Callaway R.M, Maron J.L. What have exotic plant invasions taught us over the past 20 years? Trends Ecol. Evol. 2006;21:369–374. doi: 10.1016/j.tree.2006.04.008. doi:10.1016/j.tree.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Carroll S.P, Loye J.E, Dingle H, Mathieson M, Famula T.R, Zalucki M.P. And the beak shall inherit—evolution in response to invasion. Ecol. Lett. 2005;8:944–951. doi: 10.1111/j.1461-0248.2005.00800.x. doi:10.1111/j.1461-0248.2005.00800.x [DOI] [PubMed] [Google Scholar]
- Chao A. Nonparametric-estimation of the number of classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- Cho S.C, Huang Z.Y, Green D.R, Smith D.R, Zhang J.Z. Evolution of the complementary sex-determination gene of honey bees: balancing selection and trans-species polymorphisms. Genome Res. 2006;16:1366–1375. doi: 10.1101/gr.4695306. doi:10.1101/gr.4695306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J.M, Crozier R.H. Sex determination and population biology in the Hymenoptera. Trends Ecol. Evol. 1995;10:281–286. doi: 10.1016/0169-5347(95)90011-x. doi:10.1016/0169-5347(95)90011-X [DOI] [PubMed] [Google Scholar]
- Dlugosch K.M, Parker I.M. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. doi:10.1111/j.1365-294X.2008.03663.x [DOI] [PubMed] [Google Scholar]
- Ficetola G.F, Bonin A, Miaud C. Population genetics reveals origin and number of founders in a biological invasion. Mol. Ecol. 2008;17:773–782. doi: 10.1111/j.1365-294X.2007.03622.x. [DOI] [PubMed] [Google Scholar]
- Gilchrist G.W, Lee C.E. All stressed out and nowhere to go: does evolvability limit adaptation in invasive species? Genetica. 2007;129:127–132. doi: 10.1007/s10709-006-9009-5. doi:10.1007/s10709-006-9009-5 [DOI] [PubMed] [Google Scholar]
- Giraud T, Pedersen J.S, Keller L. Evolution of supercolonies: the Argentine ants of southern Europe. Proc. Natl Acad. Sci. USA. 2002;99:6075–6079. doi: 10.1073/pnas.092694199. doi:10.1073/pnas.092694199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotzek D, Ross K.G. Genetic regulation of colony social organization in fire ants: an integrative overview. Q. Rev. Biol. 2007;82:201–226. doi: 10.1086/519965. doi:10.1086/519965 [DOI] [PubMed] [Google Scholar]
- Hedrick P.W, Gadau J, Page R.E. Genetic sex determination and extinction. Trends Ecol. Evol. 2006;21:55–57. doi: 10.1016/j.tree.2005.11.014. doi:10.1016/j.tree.2005.11.014 [DOI] [PubMed] [Google Scholar]
- Heimpel G.E, de Boer J.G. Sex determination in the Hymenoptera. Annu. Rev. Entomol. 2008;53:209–230. doi: 10.1146/annurev.ento.53.103106.093441. doi:10.1146/annurev.ento.53.103106.093441 [DOI] [PubMed] [Google Scholar]
- Hoffmeister T.S, Vet L.E.M, Biere A, Holsinger K, Filser J. Ecological and evolutionary consequences of biological invasion and habitat fragmentation. Ecosystems. 2005;8:657–667. doi:10.1007/s10021-003-0138-8 [Google Scholar]
- IUCN, The World Conservation Union 2000 100 of the world's worst invasive alien species: a selection from the global invasive species database. See http://www.iucn.org/places/medoffice/invasive_species/docs/invasive_species_booklet.pdf
- Jackson J.A, Patenaude N.J, Carroll E.L, Baker C.S. How few whales were there after whaling? Inference from contemporary mtDNA diversity. Mol. Ecol. 2008;17:236–251. doi: 10.1111/j.1365-294X.2007.03497.x. [DOI] [PubMed] [Google Scholar]
- Johnson R.N, Starks P.T. A surprising level of genetic diversity in an invasive wasp: Polistes dominulus in the northeastern United States. Annu. Entomol. Soc. Am. 2004;97:732–737. doi:10.1603/0013-8746(2004)097[0732:ASLOGD]2.0.CO;2 [Google Scholar]
- Kang M, Buckley Y.M, Lowe A.J. Testing the role of genetic factors across multiple independent invasions of the shrub Scotch broom (Cytisus scoparius) Mol. Ecol. 2007;16:4662–4673. doi: 10.1111/j.1365-294X.2007.03536.x. doi:10.1111/j.1365-294X.2007.03536.x [DOI] [PubMed] [Google Scholar]
- Knowles L.L, Futuyma D.J, Eanes W.F, Rannala B. Insight into speciation from historical demography in the phytophagous beetle genus Ophraella. Evolution. 1999;53:1846–1856. doi: 10.1111/j.1558-5646.1999.tb04567.x. doi:10.2307/2640445 [DOI] [PubMed] [Google Scholar]
- Lavergne S, Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl Acad. Sci. USA. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. doi:10.1073/pnas.0607324104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura O. Molecular genetic approaches to elucidate the ecological and evolutionary issues associated with biological invasions. Ecol. Res. 2007;22:876–883. doi:10.1007/s11284-007-0389-5 [Google Scholar]
- Morrill W.L. Production and flight of alate red imported fire ants. Environ. Entomol. 1974;3:265–271. [Google Scholar]
- Murray-McIntosh R.P, Scrimshaw B.J, Hatfield P.J, Penny D. Testing migration patterns and estimating founding population size in Polynesia by using human mtDNA sequences. Proc. Natl Acad. Sci. USA. 1998;95:9047–9052. doi: 10.1073/pnas.95.15.9047. doi:10.1073/pnas.95.15.9047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M.A.F, Pascual M, Smith K.R. Genetic variation in the spread of Drosophila subobscura from a nonequilibrium population. Evolution. 2000;54:696–703. doi: 10.1111/j.0014-3820.2000.tb00071.x. doi:10.1111/j.0014-3820.2000.tb00071.x [DOI] [PubMed] [Google Scholar]
- Pascual M, Chapuis M.P, Mestres F, Balanya J, Huey R.B, Gilchrist G.W, Serra L, Estoup A. Introduction history of Drosophila subobscura in the New World: a microsatellite-based survey using ABC methods. Mol. Ecol. 2007;16:3069–3083. doi: 10.1111/j.1365-294X.2007.03336.x. doi:10.1111/j.1365-294X.2007.03336.x [DOI] [PubMed] [Google Scholar]
- Pauchard A, Shea K. Integrating the study of non-native plant invasions across spatial scales. Biol. Invasions. 2006;8:399–413. doi:10.1007/s10530-005-6419-8 [Google Scholar]
- Pimentel D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the United States. Bioscience. 2000;50:53–65. doi:10.1641/0006-3568(2000)050[0053:EAECON]2.3.CO;2 [Google Scholar]
- Roderick G.K, Navajas M. Genes in new environments: genetics and evolution in biological control. Nat. Rev. Genet. 2003;4:889–899. doi: 10.1038/nrg1201. doi:10.1038/nrg1201 [DOI] [PubMed] [Google Scholar]
- Ross K.G, Fletcher D.J.C. Genetic origin of male diploidy in the fire ant, Solenopsis invicta (Hymenoptera: Formicidae), and its evolutionary significance. Evolution. 1985a;39:888–903. doi: 10.1111/j.1558-5646.1985.tb00430.x. doi:10.2307/2408688 [DOI] [PubMed] [Google Scholar]
- Ross K.G, Fletcher D.J.C. Comparative study of genetic and social structure in two forms of the fire ant, Solenopsis invicta (Hymenoptera: Formicidae) Behav. Ecol. Sociobiol. 1985b;17:349–356. doi:10.1007/BF00293212 [Google Scholar]
- Ross K.G, Fletcher D.J.C. Diploid male production—a significant colony mortality factor in the fire ant, Solenopsis invicta. Behav. Ecol. Sociobiol. 1986;19:283–291. doi:10.1007/BF00300643 [Google Scholar]
- Ross K.G, Keller L. Ecology and evolution of social organization: insights from fire ants and other highly eusocial insects. Annu. Rev. Ecol. Syst. 1995;26:631–656. doi:10.1146/annurev.es.26.110195.003215 [Google Scholar]
- Ross K.G, Shoemaker D.D. Species delimitation in native South American fire ants. Mol. Ecol. 2005;14:3419–3438. doi: 10.1111/j.1365-294X.2005.02661.x. doi:10.1111/j.1365-294X.2005.02661.x [DOI] [PubMed] [Google Scholar]
- Ross K.G, Vargo E.L, Keller L, Trager J.C. Effect of a founder event on variation in the genetic sex-determining system of the fire ant Solenopsis invicta. Genetics. 1993;135:843–854. doi: 10.1093/genetics/135.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K.G, Vargo E.L, Keller L. Social evolution in a new environment: the case of introduced fire ants. Proc. Natl Acad. Sci. USA. 1996;93:3021–3025. doi: 10.1073/pnas.93.7.3021. doi:10.1073/pnas.93.7.3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K.G, Krieger M.J.B, Shoemaker D.D, Vargo E.L, Keller L. Hierarchical analysis of genetic structure in native fire ant populations: results from three classes of molecular markers. Genetics. 1997;147:643–655. doi: 10.1093/genetics/147.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K.G, Shoemaker D.D, Krieger M.J.B, DeHeer C.J, Keller L. Assessing genetic structure with multiple classes of molecular markers: a case study involving the introduced fire ant Solenopsis invicta. Mol. Biol. Evol. 1999;16:525–543. doi: 10.1093/oxfordjournals.molbev.a026134. [DOI] [PubMed] [Google Scholar]
- Ross K.G, Krieger M.J.B, Shoemaker D.D. Alternative genetic foundations for a key social polymorphism in fire ants. Genetics. 2003;165:1853–1867. doi: 10.1093/genetics/165.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K.G, Krieger M.J.B, Keller L, Shoemaker D.D. Genetic variation and structure in native populations of the fire ant Solenopsis invicta: evolutionary and demographic implications. Biol. J. Linn. Soc. 2007;92:541–560. doi:10.1111/j.1095-8312.2007.00853.x [Google Scholar]
- Ruttner F. Isolated populations of honeybees in Australia. J. Apic. Res. 1976;15:97–104. [Google Scholar]
- Sakai A.K, et al. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001;32:305–332. doi:10.1146/annurev.ecolsys.32.081501.114037 [Google Scholar]
- Sax D.F, Stachowicz J.J, Gaines S.D, editors. Species invasions: insights into ecology, evolution, and biogeography. Sinauer; Sunderland, MA: 2005. [Google Scholar]
- Schlaepfer M.A, Sherman P.W, Blossey B, Runge M.C. Introduced species as evolutionary traps. Ecol. Lett. 2005;8:241–246. doi:10.1111/j.1461-0248.2005.00730.x [Google Scholar]
- Schmid-Hempel P, Schmid-Hempel R, Brunner P.C, Seeman O.D, Allen G.R. Invasion success of the bumblebee, Bombus terrestris, despite a drastic genetic bottleneck. Heredity. 2007;99:414–422. doi: 10.1038/sj.hdy.6801017. doi:10.1038/sj.hdy.6801017 [DOI] [PubMed] [Google Scholar]
- Shoemaker D.D, DeHeer C.J, Krieger M.J.B, Ross K.G. Population genetics of the invasive fire ant Solenopsis invicta (Hymenoptera: Formicidae) in the United States. Ann. Entomol. Soc. Am. 2006;99:1213–1233. doi:10.1603/0013-8746(2006)99[1213:PGOTIF]2.0.CO;2 [Google Scholar]
- Strayer D.L, Eviner V.T, Jeschke J.M, Pace M.L. Understanding the long-term effects of species invasions. Trends Ecol. Evol. 2006;21:645–651. doi: 10.1016/j.tree.2006.07.007. doi:10.1016/j.tree.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nishida T, Ohsaki N. Sequential rapid adaptation of indigenous parasitoid wasps to the invasive butterfly Pieris brassicae. Evolution. 2007;61:1791–1802. doi: 10.1111/j.1558-5646.2007.00165.x. doi:10.1111/j.1558-5646.2007.00165.x [DOI] [PubMed] [Google Scholar]
- Tschinkel W.R. Sociometry and sociogenesis of colonies of the fire ant Solenopsis invicta during one annual cycle. Ecol. Monogr. 1993;63:425–457. doi:10.2307/2937154 [Google Scholar]
- Tschinkel W.R. Harvard University Press; Cambridge, MA: 2006. The fire ants. [Google Scholar]
- Tsutsui N.D, Suarez A.V, Grosberg R.K. Genetic diversity, asymmetrical aggression, and recognition in a widespread invasive species. Proc. Natl Acad. Sci. USA. 2003;100:1078–1083. doi: 10.1073/pnas.0234412100. doi:10.1073/pnas.0234412100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugelvig L.V, Drijfhout F.P, Kronauer D.J.C, Boomsma J.J, Pedersen J.S, Cremer S. The introduction history of invasive garden ants in Europe: integrating genetic, chemical and behavioural approaches. BMC Biol. 2008;6:11. doi: 10.1186/1741-7007-6-11. doi:10.1186/1741-7007-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargo E.L, Fletcher D.J.C. Effect of queen number on the production of sexuals in natural populations of the fire ant, Solenopsis invicta. Physiol. Entomol. 1987;12:109–116. doi:10.1111/j.1365-3032.1987.tb00729.x [Google Scholar]
- Yokoyama S, Nei M. Population dynamics of sex-determining alleles in honey bees and self-incompatibility alleles in plants. Genetics. 1979;91:609–626. doi: 10.1093/genetics/91.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed A, Packer L. Complementary sex determination substantially increases extinction proneness of haplodiploid populations. Proc. Natl Acad. Sci. USA. 2005;102:10 742–10 746. doi: 10.1073/pnas.0502271102. doi:10.1073/pnas.0502271102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed A, Constantin Ş.A, Packer L. Successful biological invasion despite a severe genetic load. PLoS ONE. 2007;2:e868. doi: 10.1371/journal.pone.0000868. doi:10.1371/journal.pone.0000868 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure with likelihood values for ‘a’
Caption for electronic supplementary material figure S1