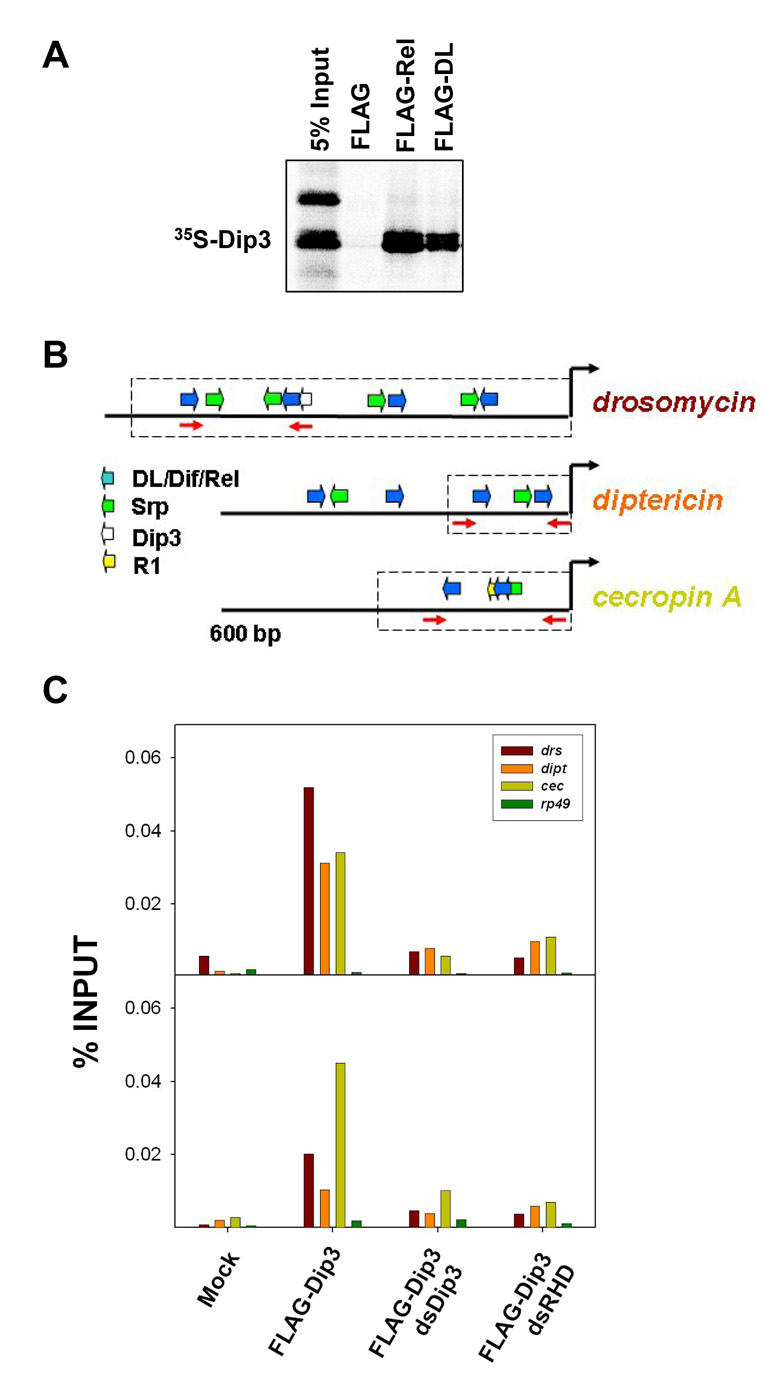
Figure 6. Dip3 is present at the enhancers of the antimicrobial defense genes.
(A) Approximately 500 ng of FLAG-DL and FLAG-Rel bound to FLAG-Beads were incubated with equivalent amounts of in vitro translated 35S-Dip3. Dip3 bound to DL and Rel but not to beads alone.
(B) The 5’ flanking regions of three antimicrobial defense genes [26], indicating binding sites for the RHD proteins DL, Dif, and Rel, and binding sites for the GATA factor Serpent (Srp). Also indicated is a potential binding site for Dip3 in drosomyocin and the Region 1 (R1) motif in cecropin A [29]. The dashed boxes indicate regions of the gene that have been experimentally confirmed [26] to direct expression patterns that mimic those of the intact genes. The primer pairs used for amplification in the ChIP assays (Figure 6B) are shown as red arrows.
(C) Chromatin immunoprecipitation assays carried out on cells expressing FLA-Gtagged Dip3, DL, His-Dif, Rel and the indicated double stranded RNAs. Quantitative real time PCR was used to quantify levels of drs, dipt, and, cecA 5’ flanking DNA in the immunoprecipitate (PCR primers are shown in A). The rp49 gene was used as a negative control. The two panels are independent repetitions of the ChIP experiment.