Abstract
We report very high gene targeting frequencies in Drosophila by direct embryo injection of mRNAs encoding specific zinc-finger nucleases (ZFNs). Both local mutagenesis via nonhomologous end joining (NHEJ) and targeted gene replacement via homologous recombination (HR) have been achieved in up to 10% of all targets at a given locus. In embryos that are wild type for DNA repair, the products are dominated by NHEJ mutations. In recipients deficient in the NHEJ component, DNA ligase IV, the majority of products arise by HR with a coinjected donor DNA, with no loss of overall efficiency in target modification. We describe the application of the ZFN injection procedure to mutagenesis by NHEJ of 2 new genes in Drosophila melanogaster: coil and pask. Pairs of novel ZFNs designed for targets within those genes led to the production of null mutations at each locus. The injection procedure is much more rapid than earlier approaches and makes possible the generation and recovery of targeted gene alterations at essentially any locus within 2 fly generations.
Keywords: coilin, DNA ligase IV, DNA repair, PAS kinase, targeted mutagenesis
The basis of genetic analysis is to explore the phenotypic consequences of alterations in DNA sequences. In forward genetics, mutations responsible for particular phenotypes are traced back to their genomic location. In reverse genetics, a genomic target is identified and mutations are directed to it. The latter approach is very powerful in elucidating gene function, particularly when any desired sequence change can be introduced. Methods to accomplish this have been devised for yeast (1), mice (2), and Drosophila (3), but the frequency of targeted mutagenesis is typically quite low, and such procedures are not available for many experimental organisms.
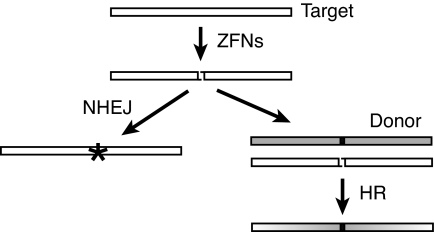
Zinc-finger nucleases (ZFNs) are proving to be powerful tools for directed genome manipulation (4, 5). The nonspecific cleavage domain of these proteins is linked to a set of DNA-binding ZFs that can be modified to recognize a wide range of DNA sequences (6, 7). Because of the requirement for dimerization of the cleavage domain (8), a pair of ZFNs is designed for any particular target (7). Upon binding to the target sequence, the dimer introduces a double-strand break (DSB) with 4-nt 5′ overhangs. Integrity of the broken chromosome is restored by cellular DNA repair functions (Fig. 1). Nonhomologous end joining (NHEJ) often produces small insertions and/or deletions at the junction, creating mutations precisely at the cleavage site (9). Cells also repair DSBs by homologous recombination (HR), and modified versions of the target gene supplied by the experimenter can serve as the repair template, thereby introducing designed modifications (10, 11).
Fig. 1.
Repair outcomes after a targeted, ZFN-induced DSB. ZFNs make a DSB in the chromosomal target, leaving a 4-base 5′ overhang. The break can be repaired by NHEJ, leading to localized mutations (star). Alternatively, a marked donor DNA can be used as a template to repair the break by HR, leading to the incorporation of specific mutant sequences (black box). Shading in the HR product indicates that sequences for some distance on either side of the break may be incorporated from the donor.
Applications of ZFNs have been reported in a number of organisms and experimental systems, including Xenopus oocytes (12), Drosophila (9, 10, 13), cultured human cells (11, 14–21), cultured plant cells (20, 22), whole plants (23), nematodes (24), and zebrafish (25, 26). For each application, delivery of the ZFNs and donor DNA must be adapted to the system at hand. In the case of Drosophila, we have achieved very high levels of targeted mutagenesis by NHEJ and gene replacement by HR based on heat shock induction of designed ZFNs (13). The procedure, however, required elaborate genetic constructions to place 2 ZFN coding sequences and the donor in the genome along with transgenes for FLP and I-SceI (which activate the donor by excision and linearization; ref. 27). We sought to simplify the procedure and make it more accessible to interested researchers.
In this study we present an embryo injection method that yields new, targeted mutations by both HR and NHEJ. The frequencies are high enough that novel mutations are recovered readily without the need for a known phenotype. We investigated several parameters of the HR process and found that best results were obtained with a circular donor DNA carrying several kilobases of homology to the target on both sides of the ZFN-induced break. Remarkably, when we used recipient embryos mutant for DNA ligase IV, a component of the canonical NHEJ pathway (28), the bias was shifted strongly toward products of HR with a coinjected donor DNA.
To demonstrate the utility of the injection method, we designed new pairs of ZFNs for targets in the Drosophila genes for coilin, a defining component of nuclear Cajal bodies (29, 30), and PAS kinase, a serine/threonine kinase that plays a role in metabolic regulation in yeast and mammals (31). The new mutations we generated will allow us to take advantage of Drosophila genetic analysis to study the detailed functions of these proteins.
Results
RNA Injections for ZFN Expression and NHEJ.
Embryo injection has been the method of choice for introducing transgenes into Drosophila for >25 years (32). In this procedure the P element transposase is expressed from an injected plasmid and leads to the hopping of a modified transposon from a separate plasmid into the genome. It seemed likely that ZFNs could also be expressed upon injection. We tried several approaches, including injection of purified proteins and DNA constructs with several different promoters. Although it may be that these formats could succeed—for example, with a different promoter—we achieved best results with injection of synthetic mRNAs (3, 33) for a pair of ZFNs.
The initial test system involved materials for the rosy (ry) gene, which gave us the highest frequencies of mutagenesis in the heat shock procedure (13). As described in Materials and Methods, the coding sequences for the ryA and ryB ZFNs were cloned into an in vitro transcription vector. RNAs synthesized from these constructs encode an N-terminal nuclear localization signal (NLS) and carry a 5′ cap and a 3′ poly(A) tail. Although an NLS was not necessary in our earlier protocol, it was essential to the success of the injection experiments.
To assess targeted mutagenesis via NHEJ, synthetic mRNAs were produced separately for the ryA and ryB ZFNs, mixed, and injected together into wild-type embryos. When adults eclosed, they were crossed to flies carrying known ry mutations to reveal new mutations generated by ZFN cleavage. Progeny of these crosses were scored for ry offspring, each of which represents an individual gamete derived from mutagenesis in the parental germ line. For both males and females, we report the percentage of fertile parents that yielded ry mutants, the average number of mutants per fertile parent, and the percentage of all offspring that were mutant. In the experiment shown in Table 1, large numbers of ry mutants were obtained from both male and female injected parents. Nearly half of the injected parents yielded mutant offspring, hundreds of mutants were recovered from those parents, and mutants represented ≈10% of all offspring.
Table 1.
NHEJ mutagenesis at the ry locus by RNA injection
| Parents | Number | Yielders, n (%) | Mutants, n | Mut/par, n | Mut/total, % |
|---|---|---|---|---|---|
| Female | 55 | 23 (42) | 356 | 6.5 | 7 |
| Male | 44 | 18 (41) | 434 | 9.9 | 13 |
The number of injected parents that produced fertile crosses is shown, along with the number (and percent) of those that yielded mutant offspring. The total number of mutants is reported, along with the mutants per fertile parent (Mut/par) and mutants as a proportion of total progeny (Mut/total).
The sequences of representative mutant ry genes were determined. They showed the usual pattern of small deletions and insertions at the ZFN cleavage site (data not shown). There were no systematic differences from the types of mutations recovered in heat shock experiments (9, 13). Individual parents typically gave multiple mutant offspring, including clusters of the same mutation (13). These sibling groups tended to contain larger clusters and fewer different sequences than similar sibling groups isolated from the heat shock experiments. This is likely because the mutations were induced at an earlier stage of development, when there were fewer germ line cells that subsequently underwent expansion en route to gamete production.
The yield of fertile adults from these injections was quite variable and often much lower than for standard P element DNA injections. With the ry ZFNs, the percentage of injected embryos that emerged as fertile adults ranged from 4% to 30%, and losses occurred at all stages of development. This may be due to consequences of injecting RNA or to the activity of the ZFNs. In other situations some toxicity of ZFNs in Drosophila has been described (9), although this was not manifested by the ryA and ryB proteins (13).
Mutagenesis of New Drosophila Genes.
Because of its readily scored mutant phenotype, the ry gene provided a useful test system. We also produced NHEJ mutations in the yellow gene by RNA injection, albeit at lower frequencies. In both cases the efficiency was high enough that new mutations would have been recoverable by molecular analysis, without reliance on a known mutant phenotype. To demonstrate this and to extend the ZFN approach to additional independent targets, we chose to attack the Drosophila genes encoding the coilin (coil, CG8710) (30) and PAS kinase (pask, CG3105) (34) proteins.
Coilin is the signature component of nuclear structures called Cajal bodies (29). We searched the coil genomic sequence for regions composed of DNA triplets for which highly specific ZFs are available (7). The particular sequence that was chosen is shown in Fig. 2. It has the standard configuration for ZFN targets: 2 sets of 3 triplets for which ZFs exist, separated by a 6-bp spacer. Five of the 6 triplets are of the form 5′-GNN-3′; fingers for such triplets are the best characterized. The sixth triplet is CTA, for which a good finger has been described (35). Notably, this is the first time we have used a non-GNN-directed finger for targeting in Drosophila. Coding sequences for the corresponding ZFNs were designed, constructed, cloned, transcribed, and injected into wild-type recipients as described in Materials and Methods.
Fig. 2.
ZFN targets in the coil and pask genes. Both strands are shown in the top lines for each target, and the triplets to which ZFs were designed are in bold, red type. Points of expected cleavage on each strand are indicated with arrowheads. The start codon for the long form of coilin is underlined. Mutations recovered from the offspring of injected parents are shown in the context of the top strand. Dashes indicate deleted residues. Substitutions and insertions are shown in bold, blue type. The first coil mutant was recovered twice independently. The 4-bp insertion in the bottom pask mutant is a fill-in and blunt join of the 4-base overhang created by ZFN cleavage.
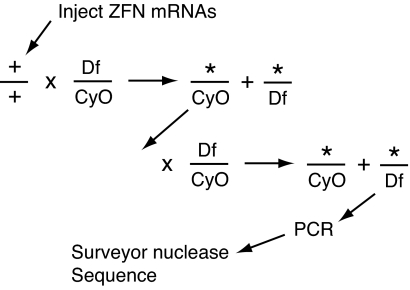
The ZFN target resides in coding sequence very near the translational start codon, so we expected NHEJ mutations to create null alleles. Because we did not know the phenotype of such mutants, we screened offspring of injected parents by molecular analysis, using a protocol that made no a priori assumption about mutant viability. Parents were crossed to partners that carried a deficiency covering the coil gene [Df(2R)CA53] opposite a second chromosome balancer (CyO) (Fig. 3). This yields offspring with candidate chromosomes either over the deficiency or over the balancer. No obvious phenotypes were observed in the former class, but the frequency of mutagenesis was not known at that stage. Candidates with the balancer were crossed again to the Df(2R)CA53/CyO strain. Because all of the candidate chromosomes were viable over the deficiency, offspring with the Df chromosome were chosen for molecular analysis.
Fig. 3.
Scheme for isolation of ZFN-induced mutations in the coil gene. mRNAs for the designed ZFNs were injected into wild-type embryos. When adults eclosed they were crossed to parents carrying a deficiency (Df) that includes the coil gene on the second chromosome and a balancer with a dominant marker (CyO). Candidate chromosomes (*) carried over the balancer were isolated individually by crossing again to the same strain. From the second cross, and in some cases from the first cross, individual alleles carried over the deficiency were subjected to molecular analysis.
A region around the coil target was amplified by PCR from genomic DNA of each of the candidates. In some cases individual amplicons were sequenced directly; in others, pools of genomic DNA from 3 nonsibling flies were amplified together and subjected to reaction with the mismatch-specific Surveyor (CelI) nuclease (36, 37). When the latter assay revealed heteroduplexes, PCR products from individual genomes were then sequenced, and several small deletions and insertions were identified (Fig. 2).
In one experiment we estimated the frequency of mutagenesis by amplifying and sequencing the coil target from 91 F1 offspring carrying the Df chromosome from 45 different injected parents. Seven of these, from 5 parents, were deletion mutants, a frequency of 8% of the tested offspring. In some of these cases the Surveyor assay was also performed, and there was good correspondence between it and the individual sequencing. In a separate experiment, a coil mutation frequency of ≈5% was obtained.
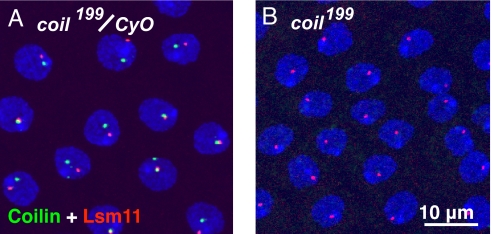
The 6 new coil mutations all created translational frameshifts very close to the start codon of the longer identified ORF (Fig. 2). In separate homozygous lines for 2 of these mutations, no coilin protein was detectable either by Western blot analysis (data not shown) or by immunocytochemistry (Fig. 4). Thus, the mutations were apparent nulls. These flies showed no reduction in viability or fertility. A more extensive analysis of the consequences of the loss of coil has been presented, including examination of ZFN-induced mutants (30).
Fig. 4.
Cells of the ejaculatory duct stained with antibodies against Drosophila coilin (green) and Drosophila Lsm11 (red). DNA is stained with DAPI (blue). (A) In flies heterozygous for coil199 and the balancer CyO, a single coilin-positive Cajal body and a single Lsm11-positive histone locus body are detectable in each nucleus. (B) In the homozygous coil199 fly, coilin is absent and only the histone locus body is detectable.
A very similar approach was taken to produce new mutations in the Drosophila gene for PAS kinase (pask). The sequence chosen for targeting is shown in Fig. 2; it also contained 1 non-GNN triplet. In a small-scale pilot study, 3 independent mutations (Fig. 2) were identified in the progeny of 1 of 5 surviving parents that were injected as embryos with the pask-directed ZFN mRNAs. The viability of flies injected with these ZFNs was considerably lower than that observed for the ry or coil enzymes. Two of the pask mutations are frameshifts that should be nulls; their phenotype is currently under examination. Loss of PAS kinase has shown rather subtle effects in other organisms (31), and the ease of manipulating its expression in Drosophila should aid in characterizing the enzyme's functions.
HR with Donor DNA by Injection.
The ability to generate new targeted mutations by ZFN cleavage and NHEJ is powerful, but one often wants to introduce specific, designed modifications into a gene of interest. In our earlier studies we induced very efficient gene replacement by generating a linear donor DNA in situ in conjunction with target cleavage by ZFNs (10, 13). It seemed straightforward to coinject a donor DNA along with the ZFN mRNAs in the new procedure. New mutant ry genes were assayed after PCR amplification for the presence of the XbaI site that replaces the ZFN recognition sequences in the donor (13) and characterized as products of NHEJ (XbaI−) or HR (XbaI+).
After testing several different formats for delivery of a homologous donor DNA for the ry locus, we found the following:
A circular donor was more effective than a linear configuration (Table 2). No HR products were recovered after coinjection of either a 4.16-kb linear fragment or a 0.5-kb PCR product with homology to both sides of the ZFN-induced break. When the same 4.16-kb donor was introduced as part of a circular plasmid, HR products represented a small but easily identified proportion of new mutations.
Extending donor homology on either side of the break had little effect on the frequency of HR (data not shown). The original 4.16-kb donor had 3.16 kb of homology on the 5′ side and 1.0 kb of homology on the 3′ side of the break (13). Two donors with additional homology were tested: the 7.46-kb “symmetrical” donor had 3.16 kb of 5′ of the ZFN target and 4.3 kb of 3′ of the target; the 7.1-kb “asymmetrical” donor had 6.1 kb of 5′ and 1.0 kb of 3′ of the target.
Coinjection of DNA seemed to decrease the overall yield of ry mutants in some cases, and the effect was more severe at higher DNA concentrations. It seems possible that large amounts of DNA bind the ZFN proteins through nonspecific interactions, thereby decreasing their effective concentrations.
Table 2.
Injections with donor DNA
| Donor | Parents, n | Yielders, n (%) | Mutants, n | HR/total, n (%) |
|---|---|---|---|---|
| 4.16 kb circ, 1.5 mg/mL | 61 | 31 (51) | 437 | 24/355 (6.8) |
| 4.16 kb, lin, 0.85 mg/mL | 157 | 63 (40) | 1380 | 0/52 (0) |
| 0.5 kb, lin, 0.125 mg/mL | 92 | 6 (6) | 30 | 0/25 (0) |
Injection mixes included the ryA and ryB mRNAs and the indicated donor DNAs and concentrations. Parents, yielders, and mutants are as in Table 1. The number of mutations that were products of HR with donor DNA is given in the final column, over the total number subjected to molecular analysis, with the percent HR in parentheses. The 3-kb plasmid backbone was present with the 4.16-kb donor in both the circular and linear configurations. The concentration of the 0.5-kb PCR product was adjusted so that the concentration of donor molecules was similar to that with the longer donor. The yield of mutants was significantly lower with the 0.5-kb donor in 2 experiments, for unknown reasons.
High Frequencies of HR in the Absence of DNA Ligase IV.
Since both HR and NHEJ are stimulated by ZFN cleavage of the target, we reasoned that the yield of HR products might be improved in a background deficient for a key NHEJ component. In fact, we found this was true in the heat shock protocol (A.B., K.J.B., J.K.T., and D.C., unpublished data). Drosophila mutants lacking DNA ligase IV (lig4−) are viable and fertile (38, 39). We injected embryos from such a strain with the ryA and ryB ZFN mRNAs along with either the 4.16-kb or 7.46-kb donor DNAs.
As shown in Table 3, the results were quite dramatic. In the lig4 background, the yield of mutants was as high as in any of our injection experiments, both in terms of parents yielding mutants and total number of induced mutations. However, essentially all of the mutants were the result of HR, with very rare NHEJ products. DSBs that are substrates for NHEJ in a wild-type background apparently become substrates for HR in the absence of DNA ligase IV, and the injected donor served as an effective template.
Table 3.
Effect of lig4 mutation
| Donor | lig4 | Parents, n | Yielders, n (%) | Mutants, n | HR/total, n (%) |
|---|---|---|---|---|---|
| 4.16 kb | + | 218 | 85 (39) | 1634 | 99/889 (11) |
| 4.16 kb | − | 37 | 17 (46) | 402 | 111/112 (99) |
| 7.46 kb | + | 120 | 32 (27) | 1048 | 57/393 (14) |
| 7.46 kb | − | 21 | 7 (33) | 227 | 66/67 (98) |
Injection mixes included ryA and ryB mRNAs and the indicated donor DNAs. Entries are as in Table 2.
The ry gene has consistently been our best ZFN target in Drosophila. The effect of eliminating lig4 was also tested for the case of the yellow gene (9, 10, 13). The overall efficiency of mutagenesis was lower at this target, but the lig4 effect was similar. Among ZFN-induced y mutants analyzed, from injections of wild-type embryos 0 of 14 were HR products, whereas 16 (70%) of 23 from lig4 embryos had incorporated the donor.
Discussion
The method we report here of delivering ZFNs to Drosophila by direct embryo injection greatly simplifies both targeted mutagenesis and targeted gene replacement with these reagents. New alleles can be identified within 2 or 3 fly generations, depending on the expected phenotype, and including a homologous donor DNA adds no time to the experiment. The previous heat shock method (13) required more generations because of extensive strain building, and addition of a donor entailed additional complexity. The injection protocol will make ZFN-directed genome modification much more accessible. The frequencies of both NHEJ and HR achieved here are sufficiently high after injection that new alleles can be isolated without the need for a detectable phenotype.
RNA injection may be additionally advantageous for at least 2 reasons. First, no ZFN-encoding transgenes are inserted into the genome, thus avoiding unintended insertional mutagenesis. Second, the injected RNAs presumably have limited duration, so the nucleases are present for a limited period. Although we do not know how long either the RNAs or the nuclease proteins persist, it seems likely the time would ultimately be shorter than in a situation where ZFN transgenes were expressed. A potential drawback is that RNA is more difficult to handle reproducibly than DNA, and we have experienced variation among batches of in vitro transcripts. Nonetheless, effective commercial kits for in vitro transcription are available.
Although both NHEJ and HR repair DSBs in the injection and heat shock procedures, we observed some differences in detail. Although a linear donor was most effective in heat shock experiments, only circular donors worked upon injection. Perhaps linear DNAs are degraded by exonucleases in the embryonic syncytial cytoplasm, whereas the donor is generated within the nucleus in the heat shock protocol. Alternatively, linear molecules may be concatenated by end-joining activities in embryos and rendered less capable of entering pole cell nuclei. In addition, the repair capabilities of embryonic and larval germ cell nuclei may be inherently different. Another difference was that an NLS on the ZFNs was absolutely required for injections, but it was not used in the heat shock experiments. We speculated earlier that the ZFNs might gain access to the genome upon nuclear membrane breakdown in dividing germ cell precursors in heat shocked larvae (13). With a shorter effective ZFN half-life after injection, such passive access may not be available to nuclei of slowly dividing embryonic pole cells.
When a donor DNA was included, the proportion of HR products was lower in injections of wild-type embryos than in heat shock experiments. This discrepancy was more than compensated by injecting into lig4 mutant embryos. The overall yield of altered targets was not decreased, but the proportion representing gene replacement by HR rose dramatically. This might be expected when the major NHEJ process is disabled. It has been observed, however, that removing DNA ligase IV does not always reduce end joining to this extent (39–42). Since lig4− flies are viable and fertile, we see no major drawbacks to using such strains as recipients for RNA and DNA injections when gene replacements are desired. Even in organisms where lig4 mutations are not well tolerated, it may be possible to enhance HR by transient depletion or inhibition of its activity.
Each time a new gene is selected for targeting, new ZFNs must be designed and constructed. We have relied on modular assembly of ZF domains from publicly available databases (7, 43). This approach has been questioned (44), and it is certainly true that more elaborate screening and selection of ZF domains has proved effective (20, 25, 26). One feature of our successful experiments is that we restricted ourselves to target sequences composed largely or exclusively of GNN triplets because these have been most extensively tested. Additional assessments of the modular design approach are clearly warranted.
For simple targeted mutagenesis in Drosophila, as we achieved at the coil and pask loci, we recommend transcribing the corresponding ZFN coding sequences in vitro and injecting the mRNAs into wild-type embryos. Screening as outlined in Fig. 3 will reveal new mutants when cleavage has been effective. For gene replacements, we recommend injecting ZFN mRNAs and the desired donor DNA into lig4 mutant embryos because the frequency of HR is very high and very few NHEJ products are produced. If ZFN toxicity is suspected based on preliminary experiments, the use of cleavage domains modified at the dimer interface to reduce homodimerization can be considered (37, 45).
Materials and Methods
Fly Stocks.
Several different stocks were used as recipients for embryo injections. Canton S and a stock carrying the third chromosome from the w1118 stock were used as wild types. The lig4 stock carried the lig4169 mutation (39). Stocks and crosses to reveal new y and ry mutations were as described (9, 13). Stocks used to reveal new coil and pask mutations were w;Df(2R)CA53/CyO (FBst0003364) and Df(2R)or-BR11, cn1bw1sp1/SM6a (FBst0001721), respectively.
Plasmid Constructions.
The construction of ZFNs for the y (9) and ry (13) targets has been described. A standard Entry vector, pENTR-NLS-G-FN, was constructed by inserting a 633-bp fragment from the GFP gene in pJH4.52 pie1his11gfp (46) between the NdeI and SpeI sites of pENTR-NLS-ZFN (47) in place of the ZF coding sequences. New coding sequences for sets of ZFs for the coil and pask gene targets were synthesized from 4 long oligonucleotides as described in ref. 7. The amino acid sequences in the vicinity of the specificity-determining residues are: coilA (GTG GAT CTA target): finger 1, QNSTLTE; finger 2, TSANLSR; and finger 3, RSDALTR; coilB (GCT GGA GTG): finger 1, RSDALTR; finger 2, QSGHLQR; and finger 3, QSSDLTR; paskA (CTG GCG GAG): finger 1, RSDNLAR; finger 2, RSDDLQR; and finger 3, RNDALTE; and paskB (GAT GTG GCT): finger 1, QSSDLTR; finger 2, RSDALTR; and finger 3, TSGNLVR. These coding sequences were cloned between the NdeI and SpeI sites of the pENTR-NLS-G-FN vector in place of the GFP fragment and confirmed by sequencing. The NLS-ZFN sequences were transferred to the in vitro transcription vector pCS2-DEST with a Clonase (Invitrogen) reaction (47). The same procedure was used to place the ryA, ryB, yA, and yB ZFN coding sequences in pCS2-DEST.
Extended ry donor DNAs were made from the original 4.16-kb version in the pBluescript vector pBSrydon4.1 (13). The 5′ extended donor was created by cutting the ApaI–EcoRI fragment from pDM30 and cloning it into pBSrydon4.1 digested with the same enzymes. This adds 2.92 kb of homology to the 5′ end of the gene, creating the 7.1-kb asymmetric donor. To produce the 3′ extended donor, a 3.3-kb PCR fragment was amplified from the ry locus of flies with the genotype Df(2R)bw-HB132, FrdHB132/SM6a, beginning at the HindIII site where the pDM30 construct ends. This was inserted by TA cloning into pGem-T (Promega). The 4.16-kb donor segment was excised as an EcoRI–HindIII fragment from pBSrydon4.1 and cloned into the pGem construct, creating the 7.46-kb symmetrical donor. A short, linear donor was produced by PCR amplification of a symmetrical 500-bp fragment from pBSrydon4.1.
RNA Injections.
Plasmid DNAs were purified with a Qiagen maxi-prep kit and concentrated by ethanol precipitation. Those for in vitro transcription were digested with NotI and repurified. They were transcribed with the AmpliScribe SP6 High Yield Transcription Kit (Epicentre) and capped afterward with the ScriptCap m7G Capping System (Epicentre). Pairs of ZFN RNAs were concentrated and mixed at concentrations ranging from 0.25 mg/mL to 1.0 mg/mL each RNA. Donor DNA was included in these mixes at concentrations from 0.125 mg/mL to 6.0 mg/mL. Best results were ultimately obtained with 0.25 mg/mL each RNA and 0.5–1.0 mg/mL DNA. Embryos were injected (200–300 in each experiment), collected, and reared by standard methods.
Analysis of Mutations.
Mutations at the y and ry loci were recovered and analyzed as described previously (9, 13). Mutations in the coil gene were recovered by crossing injected flies to partners heterozygous for a deletion that includes the entire coil gene [Df(2R)CA53/CyO]. Flies having candidate chromosomes and the CyO balancer were crossed again to the Df(2R)CA53/CyO strain. Adults lacking the balancer were analyzed for mutations. DNA was prepared from individual flies; the coil target was amplified by PCR and sequenced (47). In other cases DNA from 3 nonsibling flies was mixed and amplified, then digested with Surveyor nuclease and analyzed by gel electrophoresis (36). When mismatches were detected, DNAs from individual flies were sequenced. Mutations in pask were recovered similarly. Injected adults were crossed to Df(2R)or-BR11, cn1bw1sp1/SM6a. Progeny of fertile animals carrying the targeted chromosome over the deficiency were collected, and DNA was prepared and analyzed as above. Stocks of each mutation were established.
Immunostaining.
Testes and associated organs were removed from well-fed adult male flies in Grace medium (48) and fixed for 10 min in 4% paraformaldehyde in PBS (135 mM NaCl, 2.5 mM KCl, 4.3 mM Na2HPO4, and 1.5 mM KH2PO4, pH 7.2). All subsequent steps included 0.3% Triton X-100 in the medium. Tissues were rinsed for several hours in PBS, blocked for 1 h in 10% horse serum in PBS, and stained overnight with a mixture of antibodies against dcoilin raised in guinea pig (30) and dLsm11 raised in rabbit (49). Primary sera were used at a dilution of 1:2,000 in 10% horse serum. Tissues were then stained overnight with secondary antibodies Alexa Fluor 488 goat anti-guinea pig and Alexa Fluor 594 goat anti-rabbit (Invitrogen), and the DNA-specific dye DAPI (4′,6-diamidino-2-phenylindole), each at 1 μg/mL in 10% horse serum. Tissues were mounted in 50% glycerol plus 1 mg/mL 1,4-diaminobenzene on standard microscope slides and observed by confocal microscopy (Leica SP2). Contrast in the images was adjusted with Photoshop (Adobe Systems).
Acknowledgments.
This work was supported by National Institutes of Health Grants R01 GM078571 (to D.C.) and R01 GM33397 (to J.G.G.), and in part by a University of Utah Cancer Center support grant.
Footnotes
The authors declare no conflict of interest.
References
- 1.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 2.Capecchi MR. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 3.Rong YS, Golic KG. In: Insect Transgenesis Methods and Applications. Handler AM, James AA, editors. Boca Raton, FL: CRC Press; 2000. pp. 53–75. [Google Scholar]
- 4.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 5.Cathomen T, Joung JK. Zinc-finger nucleases: The next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 6.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to FokI cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 8.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 11.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 12.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urnov FD, et al. Highly efficient endogenous gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 15.Alwin S, et al. Custom zinc-finger nucleases for use in human cells. Mol Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 16.Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Moehle EA, et al. Targeted gene addition iinto a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombardo A, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 19.Santiago Y, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeder ML, et al. Rapid “Open-Source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright DA, et al. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- 23.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 28.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 29.Gall JG. The centennial of the Cajal body. Nature Rev Mol Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- 30.Liu JL, et al. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-05-0525. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao HX, Rutter J. The role of PAS kinase in regulating energy metabolism. IUBMB Life. 2008;60:204–209. doi: 10.1002/iub.32. [DOI] [PubMed] [Google Scholar]
- 32.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 33.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage PhiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutter J, Michnoff CH, Harper SM, Gardner KH, McKnight SL. PAS kinase: An evolutionarily conserved PAS domain-regulated serine/threonine kinase. Proc Natl Acad Sci USA. 2001;98:8991–8996. doi: 10.1073/pnas.161284798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreier B, et al. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 36.Qiu P, et al. Mutation detection using Surveyor nuclease. Biotechniques. 2004;36:702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 37.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome cleavage. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 38.Gorski MM, et al. The Drosophila melanogaster DNA ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-strand breaks and acts synergistically with Rad54. Genetics. 2003;165:1929–1941. doi: 10.1093/genetics/165.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McVey M, Radut D, Sekelsky JJ. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics. 2004;168:2067–2076. doi: 10.1534/genetics.104.033902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romeijn RJ, et al. Lig4 and Rad54 are required for repair of DNA double-strand breaks induced by P-element excision in Drosophila. Genetics. 2005;169:795–806. doi: 10.1534/genetics.104.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson-Schlitz DM, Flores C, Engels WR. Multiple-pathway analysis of double-strand break repair mutations in Drosophila. PLoS Genet. 2007;3:e50. doi: 10.1371/journal.pgen.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei DS, Rong YS. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics. 2007;177:63–77. doi: 10.1534/genetics.107.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segal DJ, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 44.Ramirez CL, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szczepek M, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 46.Strome S, et al. Spindle dynamics and the role of γ-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carroll D, Beumer KJ, Morton JJ, Bozas A, Trautman JK. In: Chromosomal Mutagenesis. Davis GD, Kayser KJ, editors. Vol 435. Totowa, NJ: Humana Press; 2008. pp. 63–77. [Google Scholar]
- 48.Grace TD. Establishment of four strains of cells from insect tissues grown in vitro. Nature. 1962;195:788–789. doi: 10.1038/195788a0. [DOI] [PubMed] [Google Scholar]
- 49.Liu JL, et al. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]