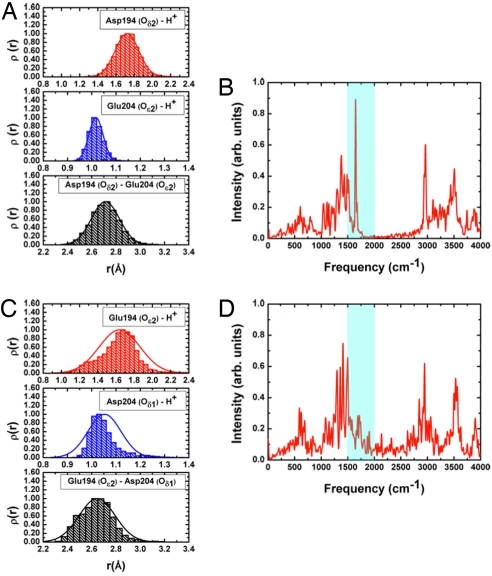
Fig. 4.
Representative results from SCC-DFTB/MM simulations for 2 mutants of bacteriorhodopsin (5 independent 2-ns trajectories each) where 1 of the key Glu residues is mutated to an aspartic. (A and B) The E194D mutant. (C and D) The E204D mutant. The distance histograms indicate that the excess proton is largely localized on Glu-204 in E194D but weakly delocalized between Glu-194/Asp-204 in E204D; this is because of the higher flexibility of Glu-194 (see main text). As a result, the continuum band in the 1,800- to 2,000-cm−1 region is absent in the computed IR spectra for E194D (B) but present in E204D (D), in agreement with the experiment (4).