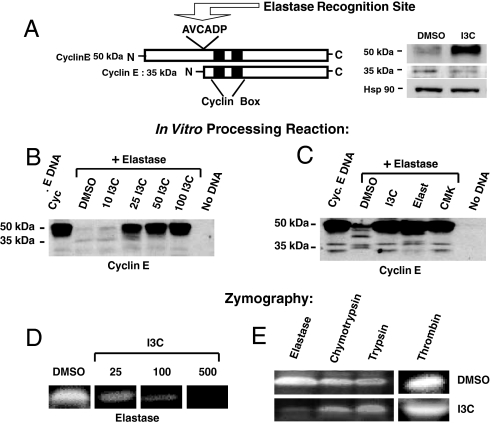
Fig. 1.
I3C inhibits elastase enzymatic activity and elastase-dependent processing of cyclin E. (A) Schematic representation of full-length cyclin E, highlighting the elastase consensus cleavage site (AVCADP). To assess endogenous cyclin protein forms, MDA-MB-231 cells were treated with or without 100 μM I3C for 48 h, and cell extracts were electrophoretically fractionated and analyzed by Western blots for the presence of the 50-kDa and hyperactive 35-kDa forms of cyclin E. (B) For the in vitro assay of elastase processing of cyclin E protein, full-length cyclin E was transcribed and translated in vitro followed by digestion of the cyclin E protein product by human neutrophil elastase in the presence of the indicated concentrations of I3C or DMSO. The “No DNA” sample was a reaction mixture without added cyclin E cDNA. Western blot analysis of the entire reaction mixtures was performed by using cyclin E-specific antibodies. (C) The in vitro elastase processing of cyclin E was assessed in the presence of the DMSO vehicle control, 50 μM I3C, and the known elastase inhibitors 25 μM elastatinal or 5 μM methoxysuccinyl-Ala-Ala-Pro-Val-chloromethylketone (CMK). Western blot analysis of the entire reaction mixture was performed by using cyclin E-specific antibodies. (D) For zymography, equal amounts of purified human elastase were electrophoretically resolved on a 10% polyacrylamide gel containing elastin as a substrate. After renaturation with 2.5% Triton X-100, each lane was cut out and placed in developing buffer containing either DMSO or the indicated concentrations of I3C for 25 h at 37 °C. Gel slices were stained with Coomassie blue followed by an overnight destain. Clear bands represent proteolytic degradation of substrates. (E) Purified human elastase, chymotrypsin, trypsin, or thrombin were electrophoretically resolved in gels containing gelatin (for evaluation of elastase, chymotrypsin, or trypsin) or fibronectin (for evaluation of thrombin), the enzymes were renatured in 2.5% Triton X-100, and individual gel slabs were developed in either DMSO or 500 μM I3C for 25 h at 37 °C.