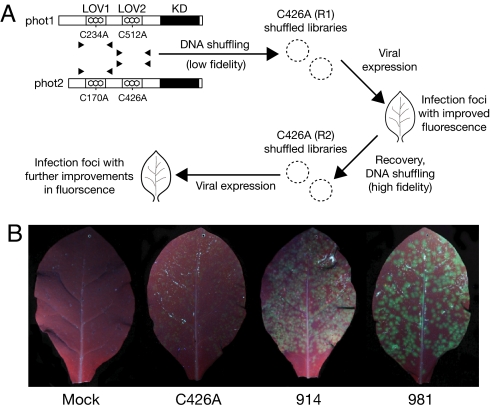
Fig. 1.
DNA shuffling of phototropin LOV domains. (A) Schematic representation of the shuffling procedure. Arabidopsis phot1 and phot2 consist of a C-terminal serine/threonine kinase domain (KD) and two photosensory LOV domains (LOV1 and LOV2) that bind the chromophore FMN. The conserved cysteine required for LOV-domain photochemistry was replaced with alanine by site-directed mutagenesis before DNA shuffling. Two sequential rounds of DNA shuffling were carried out (R1 and R2, respectively). In R1, shuffled populations were generated by using low fidelity PCR conditions. For R2, high fidelity PCR conditions were used. In each case, the LOV2 domain of Arabidopsis phot2 (C426A) was used as a template scaffold for reassembly. Shuffled populations were subjected to TMV-based expression in tobacco and screened for improved fluorescence under UV light. (B) TMV-based expression of LOV variants in leaves of Nicotiana tabacum. Images were recorded simultaneously under UV illumination to allow direct comparison of green fluorescence. Leaves were either mock inoculated or inoculated with TMV vector expressing the progenitor C426A or the brightest variants from R1 and R2 (914 and 981, respectively) and photographed 3 days post inoculation.