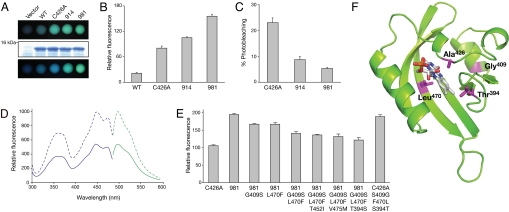
Fig. 2.
Photochemical characterization of shuffled LOV variants expressed in E. coli. (A) In vivo fluorescence in E. coli liquid cultures expressing wild-type Arabidopsis phot2 LOV2 (WT), derivative C426A and shuffled variants 914 and 981 viewed immediately under UV light (Top) or after several minutes of UV irradiation (Bottom). Equal protein levels in E. coli cultures are shown by SDS/PAGE and Coomassie Blue staining using cells transformed with the expression vector only as a control (Middle). (B) Quantification of LOV-mediated in vivo fluorescence in E. coli liquid cultures. Fluorescence intensities of liquid cultures were recorded at 495 nm upon excitation with blue light (450 nm). (C) Fluorescence loss in LOV-expressing E. coli cultures after xenon arc lamp illumination. Fluorescence intensities were recorded as in (B). (D) Fluorescence excitation and emission spectra of purified C426A (solid line) and variant 981 (dashed line). Fluorescence excitation spectra (blue) were recorded by using an emission wavelength of 495 nm, whereas fluorescence emission spectra (green) were recorded by using an excitation wavelength of 450 nm. (E) Reverse mutagenesis and quantification of 981-mediated in vivo fluorescence in E. coli liquid cultures. Point mutations indicated were introduced into 981 and the effect on in vivo fluorescence was assessed as in (B). Selective point mutations were then introduced into the progenitor C426A to confirm their role in enhancing fluorescence emission. (F) Structure of 981 was obtained by homology modeling with the program Swiss Model using the protein structure of Adiantum-capillus-veneris neochrome LOV2 (PDB entry IG28) and visualized by using PyMOL. Amino acid residues contributing to the enhanced fluorescence of 981 are indicated in magenta.